Evaluation of genetic diversity in a natural rosewood population (Dalbergia nigra Vell. Allemão ex Benth.) using RAPD markers F.S. Juchum, J.B. Leal, L.M. Santos, M.P. Almeida, D. Ahnert
and R.X. Corrêa Genet. Mol. Res. 6 (3): 543-553 (2007)
ABSTRACT. Dalbergia nigra (rosewood) is a long-lived leguminous species, which is endemic to the Brazilian Atlantic forest. Because of the high economic value of its wood, this species has been over-explored in recent years. Currently, rosewood is included in the IUCN Red List as vulnerable. We examined the genetic diversity of 87 specimens of D. nigra sampled from a continuous forest in the Veracel Reserve and Brazilwood Ecological Station, Porto Seguro, Bahia state, with random amplified polymorphic DNA markers. Grouping analyses were done using unweighted pair group method with arithmetic averages. Using the 16 most informative primers, 112 markers were obtained; 39% (44 bands) were polymorphic. A genetic similarity matrix was made based on the polymorphic bands. The dispersion graph and dendrogram analyses showed three distinct sub-populations. The degree of polymorphism was high, near that of other populations of similar species; however, it was considered low for the conservation of this species. Key words: Conservation, Leguminosae, Molecular variation, Polymerase chain reaction INTRODUCTION Dalbergia nigra Vell. Allemão ex Benth. (rosewood or Brazilian rosewood) is a tree species endemic to the Atlantic forest of Brazil; the natural distribution is in the states of Rio de Janeiro, Espírito Santo, Minas Gerais, and southern Bahia (Leão and Vinha, 1975). The Atlantic coast forest originally stretched along the coast from Rio Grande do Norte to Rio Grande do Sul; currently it is limited to less than 10% of its original area (MMA, 2000; Câmara, 2005). In addition to its ecological importance, D. nigra has a high-quality wood, which is used for the manufacturing of furniture, musical instruments and decorative objects (Almeida, 2001). According to Rizzini and Mattos Filho (1986) and Sperândio and Fonseca (1986), it is one of the most important and valuable Brazilian woods. Because of its high commercial value, rosewood has been over-explored, causing its near extinction (Sperândio and Fonseca, 1986). Rosewood is considered by the IUCN Red List as vulnerable, with a high risk of extinction in the wild in the medium-term future (Varty, 1998). Currently, a few individuals can still be found in the remaining forest fragments of the southern Bahian Atlantic forest (Almeida, 2001). Information on genetic variability is important for species that are rare in native forests and for which there is little demographic information. Genetic variability or genetic polymorphism is hereditary variation in DNA sequences among individuals of a population (Alberts et al., 1997). Differences of this nature can be found between geographically distinct populations within a local group, or within a progeny (Falconer, 1989). According to Lacy (1988) and Fritsch and Rieseberg (1996), Conservation Genetics attempts to estimate the level and distribution of the genetic variation in species that are at risk of extinction. Accurate estimates of species diversity are useful to optimize sampling strategies and for the conservation and administration of genetic resources (Hamrick et al., 1991; Schaal et al., 1991; Chalmers et al., 1992). The loss of genetic variability can be a consequence of fragmentation. Forest fragmentation is a disruptive process that cuts off and decreases gene flow, and consequently limits species evolution (Barrett and Kohn, 1991). Genetic diversity can be evaluated using genetically inherited markers that distinguish individuals or populations. Generally, genetic markers are grouped into morphologic, biochemists and molecular groups, each with specific advantages and disadvantages (Gepts, 1995). Studies of genetic variability can determine the level of genetic variation of populations or species and its distribution among populations (Hamrick, 1983). In genetic polymorphism evaluation studies, the use of molecular markers within plant species has been greatly expanded through methods that use polymerase chain reaction. random amplified polymorphic DNA (RAPD) markers provide a larger number of potential markers that are useful for the analysis of genetic diversity, often using fast, simple and reliable protocols that minimize the amount and quality of DNA required. It has been especially used to study the genetic structure of populations (Almeida, 2001) and for conservation studies of endangered organisms (Calero et al., 1999). Genetic variability analyses using RAPD have many practical applications in crop breeding and species conservation, such as: genotype classification in genetically similar groups, genitor selection, based on genetic relationship information; patent protection for new breeding varieties and the organization of germplasm banks by groups of interest (Barbosa Neto and Bered, 1998). Bai et al. (1998) verified the efficiency of RAPD markers in the detection of kindred degree between bean genotypes. Additionally, RAPD analysis has been used to describe population structure and genetic polymorphism in many species, especially in those that do not possess specific molecular tools, such as SSR, e.g., Gliricidia sepium (Chalmers et al., 1992), Buchloë dactyloides (Huff et al., 1993), Hordeum spontaneum (Dawson et al., 1993), Theobroma cacao (Russell et al., 1993), Pseudotsuga menziesii (Aagaard et al., 1995), Eucalyptus globulus (Nesbitt et al., 1995), Grevillea scapigena (Rossetto et al., 1995), Spartina alterniflora (Stiller and Denton, 1995), Populus tremuloides (Yeh et al., 1995), Argyroxiphium sandwicense (Friar et al., 1996), Medicago truncatula (Bonnin et al., 1996), Allium aaseae (Smith and Pham, 1996), Amentotaxus formosana (Wang et al., 1996), Caesalpinia echinata (Cardoso et al., 1998), Fitzroya cupressoides (Allnutt et al., 1999), Sophora toromiro (Maunder et al., 1999), Eucalyptus microtheca (Chunyang, 2000), Leucopogon obtectus (Zawko et al., 2001), Medicago truncatula (Bonnin et al., 2001), Plathymenia reticulata (Lacerda et al., 2001), Dalea purpurea (Gustafson et al., 2002), Borderea chouardii (Segarra-Moragues et al., 2005), Vigna unguiculata (Xavier et al., 2005), and some legumes (Jena et al., 2004). The preservation of Brazil’s Atlantic Forest remnant species is important for scientific and economic reasons. More information on the history, biology, ecological characteristics, and genetic bases of these species is also required, especially for endemic species, species with restricted distribution, rare species, and those in risk of extinction. Furthermore, the study of genetic diversity is important because of its evolutionary implications and the facilitation of species conservation. We evaluated the genetic diversity of a continuous population using RAPD markers. The results should help identify the best individuals for incorporation in reforestation and enrichment programs and in the formation of germplasm banks. MATERIALS ANS METHODS Population studied The study sites included natural populations from 6800 continuous hectares of forest located in the Veracel Reserve (RPPN) and in the Brazilwood Ecological Station, Porto Seguro, Bahia state (Figure 1). The D. nigra trees in this forest were mapped, and 87 of these were sampled. Leaf samples were collected from each individual and stored at -80°C until DNA extraction. Molecular analysis The DNA was extracted from the frozen leaves following the CTAB protocol (Doyle and Doyle, 1987), modified by Almeida (2001). The RAPD (Williams et al., 1990) amplifications of each individual followed the procedures described by Corrêa et al. (1999). 
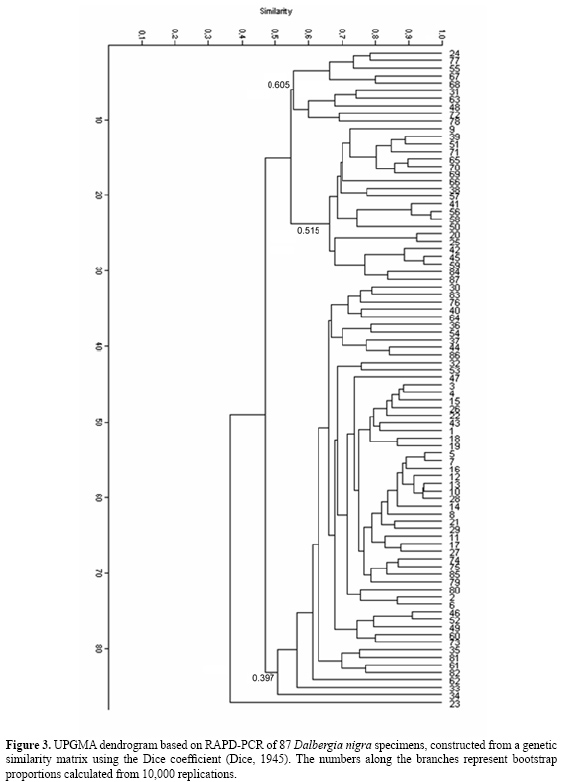
Twenty-two primers were evaluated to test their degree of polymorphism. The 16 most polymorphic primers were selected for the genetic variability analysis. DNA fragments, generated by amplification, were separated according to size on 1.2% agarose gels that were stained with ethidium bromide. These were observed using UV light and photographed with the EDAS 290 (Kodak). Reproducibility and repeatability of the amplification products were tested for each of the primers. Statistical analysis Although the visualizations varied in size, the results from each RAPD product were assumed to represent a single locus, and data were scored as the presence (1) or absence (0) of a DNA band. Only those fragments that amplified consistently were considered for analysis. The parameters and the percentage of polymorphic loci were calculated using the Past 1.34 program (Hammer et al., 2001). A matrix of genetic similarity was obtained between pairs of individuals using the Dice coefficient (Dice, 1945). The same program was used to construct a dendrogram, using unweighted pair-group method with arithmetic averages (UPGMA), which shows the relationships between individuals. A co-phenetic value was produced from the tree matrix using the COPH program from NT-SYS 1.70 software (Rohlf, 1992). RESULTS AND DISCUSSION The 16 most informative RAPD primers were used on the 87 D. nigra samples, originally tested with 22 primers, producing 112 reproducible RAPD markers, 39% were polymorphic, with an average of seven bands per primer (see Table 1). Primers OPB11, OPD11 and OPD10 gave the highest degree of variability per locus, the first two having 78% and the last having 75% polymorphic bands. The markers OPC02 and OPD03 gave low polymorphism indexes (14%). We found that D. nigra has a degree of variability within the range of that found in populations of Eucalyptus urophylla (Pigato and Lopes, 2001), Bauhinia forficate (Santos, 1994) and Myrciaria floribunda (Vasconcelos, 2002), with 23.3, 45.1 and 38.6% polymorphic bands, respectively. Based on alloenzymes, Hamrick and Godt (1990) found population level variability averages of 14.9% for tree species, 15.9% for widely distributed species, 10.9% for tropical species, and 12.4% for species pollinated by animals. Though the degree of polymorphism in D. nigra was high in comparison to figures found in these RAPD and isoenzyme studies, it was low in terms of what is desirable for species conservation. According to Kageyama et al. (2003), there is higher genetic variability within than among tree populations. Consequently, we are concerned about the conservation of rosewood. The 16 primers allowed us to discriminate the 87 specimens. Good results have been obtained for other species using a similar number of markers. For example, Dwivedi et al. (2001) evaluated the genetic diversity of 26 peanut plants with eight polymorphic markers. They determined five genotype groups and indicated that a relatively small number of markers are capable of generating information about genetic variability. Additionally, Chen et al. (1997) found 16 polymorphic markers among 40 tested, in 11 individuals, in an evaluation of soybean polymorphism. 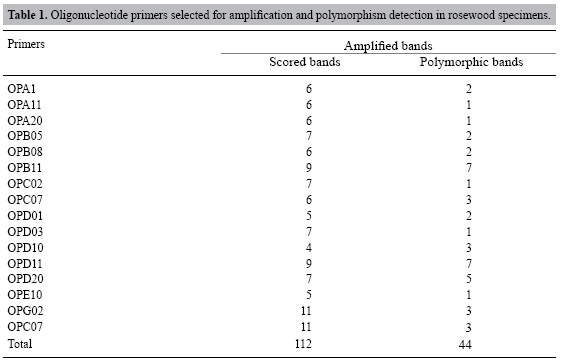
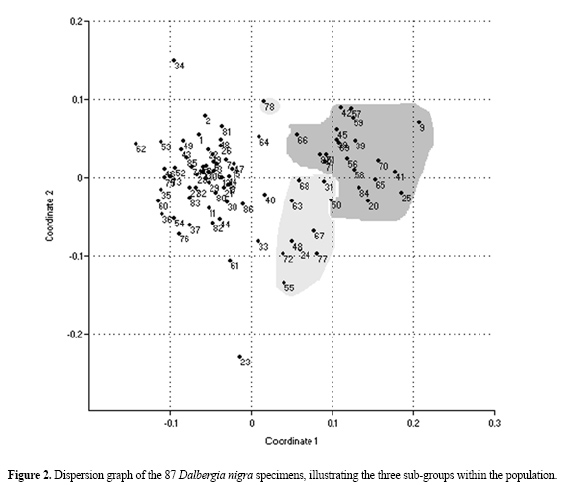
Based on the similarity analyses and distance index, similarity coefficients ranged from 0.070 to 0.965 among the 87 rosewood plants. The most similar individuals were individuals 56 and 58, with 96.5% similarity. Individuals 9 and 85 were the most distinct, with 7.7% similarity (Figures 2 and 3). Further study of factors that can explain or contribute to the analysis of the degree of polymorphism is important for the conservation of populations in isolated reserve areas. Isolation of populations can influence genetic drift and can limit intra-specific diversity. Other influential factors include the extraction of non-lumber products and fragmentation. Fragmentation disrupts the natural continuity of habitats and may cause a decrease in gene flow between populations. This causes a loss of genetic variability and limits species evolution (Barrett and Kohn, 1991). Hamrick and Murawski (1991) compared the average variability of 16 rare tree species with 16 common tree species and observed significantly higher values for the common ones. According to Ribeiro (2002), the effect of fragmentation in D. nigra populations in three forest fragments was a decrease in allele frequency and loss of alleles in four of five loci.
In this rosewood population, consisting of 87 individuals within a continuous forest, three sub-groups were distinguished. There was some intersection among individuals in the sub-groups, based on dispersion graph analysis (Figure 2). Based on the dispersion data, we calculated a similarity coefficient of 0.53 in the dendrogram (Figure 3). This may mean that there are rosewood sub-populations within this area. It would be interesting to determine if there is any relationship between the genotypic similarities and their respective phenotypes, using morphological analyses. According to Hamrick and Godt (1990), tropical tree species have their species life history and ecological characteristics reflected in intra- and inter-populational variation and in population variability. However, molecular markers do not have a direct effect on plant phenotype, and are therefore not directly affected by the selection process (Vera et al., 1999). The UPGMA dendrogram tree (Figure 3) has a co-phenetic correlation value of 0.803. Intent matrix selection suggests a cut-off at 0.65 within the dendrogram, defining at least 10 groups for future conservation applications. According to Kageyama et al. (2003), species sampling using genetic markers can help advance genetic understanding of natural tree species populations, as well as help in monitoring actions, degraded area restoration and the determination of the number of maternal trees necessary for seed collection. In addition, selection of maternal trees with high genetic variability could be used to orient seed collection, contributing to the success of restoration projects. When these issues are better understood, more accurate decisions about the future recovery and conservation of this species can be made. Molecular analysis can help us understanding genetic systems in endangered plants and contribute to existing knowledge concerning conservation of genetic resources in rare populations of D. nigra. ACKNOWLEDGMENTS Research supported by FAPESB (Fundação de Amparo à Pesquisa no Estado da Bahia, Bahia, Brazil). F.S. Juchum was the recipient of a FAPESB fellowship, and J.B. Leal and L.M. Santos were recipients of UESC fellowships. We thank Sérgio C. Coutinho for marking the trees and Vicente Militão for collecting plant material for this study. REFERENCES Aagaard JE, Vollmer SS, Sorensen FC and Strauss SH (1995). Mitochondrial DNA products among RAPD profiles are frequent and strongly differentiated between races of Douglas-fir. Mol. Ecol. 4: 441-446. Alberts B, Bray D, Lewis JR, Raff M, et al. (1997). Biologia molecular da célula. 3ª ed. Artes Médicas, Porto Alegre. Allnutt TR, Newton AC, Lara A, Premoli A, et al. (1999). Genetic variation in Fitzroya cupressoides (alerce), a threatened South American conifer. Mol. Ecol. 8: 975-987. Almeida MP (2001). Avaliação da diversidade genética de acessos ex situ de jacarandá-da-bahia (Dalbergia nigra Vell. Allemão ex Benth.) por meio de marcadores RAPD como subsídio para sua conservação. M.Sc. thesis, Universidade Estadual de Santa Cruz, Santa Cruz. Bai Y, Michals TE and Pauls P (1998). Determination of genetic relationships among Phaseolus vulgaris populations in a conical cross from RAPD markers analyses. Mol. Breed. Dordrecht 4: 395-406. Barbosa Neto JF and Bered F (1998). Marcadores moleculares e diversidade genética no melhoramento de plantas. In: Marcadores moleculares em plantas (Milack S, ed.). UFRGS, Porto Alegre, 29-41. Barrett SCH and Kohn JR (1991). Genetic and evolutionary consequences of small populations size in plants: implications for conservation. In: Genetic and conservation of rare plants (Falk DA and Holsinger KE, eds.). Oxford University Press, New York, 3-30. Bonnin I, Huguet T, Gherardi M, Properi JM, et al. (1996). High level of polymorphism and spatial structure in a selfing plant species, Medicago trunculata (Leguminosae), shown using RAPD markers. Am. J. Bot. 83: 843-855. Bonnin I, Ronfort J, Wozniak F and Olivieri I (2001). Spatial effects and rare outcrossing events in Medicago truncatula (Fabaceae). Mol. Ecol. 10: 1371-1383. Calero C, Ibáñez O, Mayol M and Rosselló JA (1999). Random amplified polymorphic DNA (RAPD) markers detect a single phenotype in Lysimachia minoricensis J.J. Rodr. (Primulaceae), a wild extinct plant. Mol. Ecol. 8: 2133-2136. Câmara IG (2005). Breve história da conservação da Mata Atlântica. In: Mata Atlântica: biodiversidade, ameaças e perspectivas. São Paulo: Fundação SOS Mata Atlântica (Galindo-Leal C and Câmara IG, eds.). Conservação Internacional, Belo Horizonte, 31-42. Cardoso MA, Provan J, Powell W, Ferreira PCG, et al. (1998). High genetic differentiation among remnant populations of the endangered Caesalpinia echinata Lam. (Leguminosae-Caesalpinioideae). Mol. Ecol. 7: 601-608. Chalmers KJ, Waugh R, Sprent JI, Simons AJ, et al. (1992). Detection of genetic variation between and within populations of Gliricidia sepium and G. maculata using RAPD markers. Heredity 69: 465-472. Chen LFO, Kuo HY, Chen MH, Lai KN, et al. (1997). Reproducibility of the differential amplification between leaf and root DNA in soybean revealed by RAPD markers. Theor. Appl. Genet. 95: 1033-1043. Chunyang L (2000). RAPD analysis of genetic variation in Eucalyptus microtheca F. Muell. populations. Hereditas 132: 151-156. Corrêa RX, Abdelnoor RV, Faleiro FG, Cruz CD, et al. (1999). Genetic distances in soybean based on RAPD markers. Bragantia 58: 15-22. Dawson IK, Chalmers KJ, Waugh R and Powell W (1993). Detection and analysis of genetic variation in Hordeum spontaneum populations from Israel using RAPD markers. Mol. Ecol. 2: 151-159. Dice LR (1945). Measures of the amount of ecologic association between species. Ecology 26: 297-302. Doyle JJ and Doyle JL (1987). A rapid DNA isolation from small amount of fresh leaf tissue. Phytochem. Bull. 19: 11-15. Dwivedi SL, Gurtu S, Chandra S, Yuejin W, et al. (2001). Assessment of genetic diversity among selected groundnut germplasm: 1- RAPD analysis. Plant Breed. 120: 345-359. Falconer DS (1989). Introduction to quantitative genetics. 3rd edn. Longman Scientific and Technical, New York. Friar EA, Robichau RH and Mount DW (1996). Molecular genetic variation following a population crash in the endangered Mauna Kea silverwood, Argyroxiphium sandwicense spp. sandwicense (Asteraceae). Mol. Ecol. 5: 687-691. Fritsch P and Rieseberg LH (1996). The use of random amplified polymorphic DNA (RAPD). In: Molecular genetic approaches in conservation (Smith TB and Wayne RK, eds.). Oxford University Press, New York, 54-73. Gepts P (1995). Genetic markers and core collections. In: Core collections of plant genetic resources (Hodgkin T, Brown AHD and Van Hintum TJL, eds.). John Wiley and Sons, Chichester, 127-146. Gustafson DJ, Gibson DJ and Nickrent DL (2002). Genetic diversity and competitive abilities of Dalea purpurea (FABACEAE) from remnant and restored grasslands. Int. J. Plant Sci. 163: 979-990. Hammer O, Harper DAT and Ryan PD (2001). PAST: paleontological statistics software package for education and data analysis. Paleontol. Electronica 4: 9. Hamrick JL (1983). The distribution of genetic variation within and among natural plant populations. In: Genetics and conservation: a reference for managing wild animal and plant populations (Shonewald-Cox CM, Chambers SM, MacBryde B and Thomas L, eds.). Benjamin Cummings, Menlo Park, 335-348. Hamrick JL and Godt MJW (1990). Allozyme diversity in plant species. In: Plant population genetics, breeding and genetic resources (Brown AHD, Clegg MT, Kahler AL and Weir BS, eds.). Sinauer Associates, Sunderland, 145-162. Hamrick JL and Murawski DA (1991). Levels of allozyme diversity in populations of uncommon Neotropical tree species. J. Trop. Ecol. 7: 395-399. Hamrick JL, Godt MJW, Murawski DA and Loveless MD (1991). Correlations between species traits and allozyme diversity: in implications for conservation biology. In: Genetic and conservation of rare plants (Falk DA and Holsinger KE, eds.). Oxford University Press, New York, 75-86. Huff DR, Peakall R and Smouse PE (1993). RAPD variation within and among natural populations of outcrossing buffalograss (Buchloë dactyloides (Nutt.) Engelm.). Theor. Appl. Genet. 86: 927-934. Jena S, Sahoo P, Mohanty S and Das AB (2004). Identification of RAPD markers, in situ DNA content and structural chromosomal diversity in some legumes of the mangrove flora of Orissa. Genetica 122: 217-226. Kageyama PY, Sebbenn AM, Ribas LA and Gandara FB (2003). Diversidade genética em espécies arbóreas tropicais de diferentes estágios sucessionais por marcadores genéticos. Sci. Forestalis 64: 93-107. Lacerda DR, Acedo MD, Filho JP and Lovato MB (2001). Genetic diversity and structure of natural populations of Plathymenia reticulata (Mimosoideae), a tropical tree from the Brazilian Cerrado. Mol. Ecol. 10: 1143-1152. Lacy RC (1988). A report on population genetics in conservation. Conserv. Biol. 2: 245-247. Leão AC and Vinha SG (1975). Ocorrência de jacarandá no Sul da Bahia. Cacau Atual. 12: 22-27. Maunder M, Culham A, Bordeu A, Allainguinllaume J, et al. (1999). Genetic diversity and pedigree for Sophora toromiro (Leguminosae): a tree extinct in the wild. Mol. Ecol. 8: 725-738. MMA - Ministério do Meio Ambiente (2000). Avaliação e ações prioritárias para a conservação da biodiversidade da Mata Atlântica e Campos Sulinos. MMA, Brasília. Nesbitt KA, Potts BM, Vaillancourt RE, West AK, et al. (1995). Partitioning and distribution of RAPD variation in a forest tree species, Eucalyptus globulus (Myrtaceae). Heredity 74: 628-637. Pigato SMPC and Lopes CR (2001). Avaliação da variabilidade genética em quatro gerações de Eucalyptus urophylla S.T. Blake por meio do marcador molecular RAPD. Sci. Forestalis 60: 119-133. Ribeiro RA (2002). Efeitos da fragmentação de habitats na estrutura genética de Dalbergia nigra (jacarandá-da-bahia): uma espécie ameaçada da Mata Atlântica. M.Sc. thesis, Universidade Federal de Minas Gerais, Belo Horizonte. Rizzini CT and Mattos Filho A (1986). Árvores e madeiras úteis do Brasil. Manual de Dendrologia Brasileira. Edgard Blucher, São Paulo. Rohlf FJ (1992). NTSYS: numerical taxonomy and multivariate analysis system. Version 1.7. Applied Biostatistic Inc., New York. Rossetto M, Weaver PK and Dixon KW (1995). Use of RAPD analysis in devising conservation strategies for the rare and endangered Grevillea scapigera (Proteaceae). Mol. Ecol. 4: 321-329. Russell JR, Hosein F, Johnson E, Waugh R, et al. (1993). Genetic differentiation of cocoa (Theobroma cacao L.) populations revealed by RAPD analysis. Mol. Ecol. 2: 89-97. Santos EMG (1994). Ecologia da polinização, fluxo de pólen e taxa de cruzamento em Bauhinia forficata Link (Caesalpiniaceae). M.Sc. thesis, Escola Superior de Agricultura "Luiz de Queiroz", Universidade de São Paulo, Piracicaba. Schaal BA, Leverich WJ and Rogstad SH (1991). A comparison of methods for assessing genetic variation in plant conservation biology. In: Genetic and conservation of rare plants (Falk DA and Holsinger KE, eds.). Oxford University Press, New York, 123-134. Segarra-Moragues JG, Palop-Esteban M, González-Candelas F and Catalán P (2005). On the verge of extinction: genetics of the critically endangered Iberian plant species, Borderea chouardii (Dioscoreaceae) and implications for conservation management. Mol. Ecol. 14: 969-982. Smith JF and Pham TV (1996). Genetic diversity of the narrow endemic Allium aaseae (Alliaceae). Am. J. Bot. 83: 717-726. Sperândio JP and Fonseca CEL (1986). Comportamento do jacarandá-da-Bahia em plantios experimentais em Manaus. In: Anais do 1º Simpósio do Trópico Úmido, Belém. Stiller JF and Denton AL (1995). One hundred years of Spartina alterniflora (Poaceae) in Willapa Bay, Washington: random amplified polymorphic DNA analysis of an invasive population. Mol. Ecol. 4: 355-365. Varty N (1998). Dalbergia nigra. In: IUCN. 2006. 2006 IUCN Red List of Threatened Species. http://www.iucnredlist.org. Accessed July 2006. Vasconcelos GMP (2002). Diversidade genética de Myrciaria floribunda (West ex Willdenow) Berg (Cambuí) em paisagem fragmentada da Serra da Mantiqueira, MG. M.Sc. thesis, Escola Superior de Agricultura "Luiz de Queiroz", Universidade de São Paulo, Piracicaba. Vera CM, Paredes MC and Becerra VV (1999). Estúdio comparativo de diversidad morfológica, isoenzimatica y RAPDs dentro y entre clases comerciales de fréjol chileno (Phaseolus vulgaris L.). Agric. Téc. 59: 247-259. Wang CT, Wang WY, Chiang CH, Wang YN, et al. (1996). Low genetic variation in Amentotaxus formosana Li revealed by isozyme analysis and random amplified polymorphic DNA markers. Heredity 77: 388-395. Williams JG, Kubelik AR, Livak KJ, Rafalski JA, et al. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18: 6531-6535. Xavier GR, Martins LMV, Rumjanek NG and Filho FRF (2005). Variabilidade genética em acessos de caupi analisada por meio de marcadores RAPD. Pesqui. Agropecu. Bras. 40: 353-359. Yeh FC, Chong DK and Yang RC (1995). RAPD variation within and among natural populations of trembling aspen (Populus tremuloides Michx.) from Alberta. J. Hered. 86: 454-460. Zawko G, Krauss SL, Dixon KW and Sivasithamparam K (2001). Conservation genetics of the rare and endangered Leucopogon obtectus (Ericaceae). Mol. Ecol. 10: 2389-2396. |
|