
ABSTRACT. Expressed sequenced tags (ESTs) were prepared to establish a baseline for molecular genetic studies of the tarnished plant bug, Lygus lineolaris (Palisot de Beauvois). The largest class of identifiable ESTs (15.2%) was from genes involved in cellular metabolic functions, including physiological processes. Twenty-seven ESTs (9.8%) were from genes associated with transcription and translation, including ribosomal genes. One hundred and forty-two of the 276 unique ESTs were from genes not previously identified from any organism. Twelve sequences appear to be associated with feeding and digestion and may be targets for pest control studies. Key words: Expressed sequence tags, Tarnished plant bug, Feeding, Lygus lineolaris, Digestion INTRODUCTION
Lygus lineolaris (Palisot de Beauvois) (Hemiptera: Miridae) is a pest of many agricultural crops, often damaging young fruit and growing stems by injecting salivary enzymes into plant tissue (Wheeler, 2001). In the cotton-growing regions of North America, insect damage to crops has changed dramatically over the past 10 years. While damage from lepidopterous pests has decreased due to the adoption of insect-resistant transgenic varieties of corn, soybean, and cotton, L. lineolaris has emerged as a leading pest insect in cotton, based on loss estimates (Williams, 2005). An understanding of the genetics associated with feeding and digestion processes in L. lineolaris is vital to development of genetic strategies to curtail losses caused by this insect. Because the genome of L. lineolaris is not currently planned for sequencing, cDNA expressed sequenced tag (EST) sequences from this insect present the most basic functional genetic information available in the short term. A small expression library was constructed, sequenced and analyzed to obtain a basic list of gene sequences from L. lineolaris. Expressed sequences that correspond to insect structural genes, digestive enzymes, and common housekeeping gene sequences were segregated and compared to other publicly available arthropod sequences. MATERIAL AND METHODS Lygus lineolaris colony was established from field collections made near Starkville, MS, USA, and originally maintained at the Biological Control & Mass Rearing Research Unit, Starkville, MS. Insects used for library construction were reared at the USDA National Biological Control Laboratory in Stoneville, MS. They were maintained on a 16-h light/8-h dark cycle, at a temperature of 24° and 20°C, respectively, and at 70% relative humidity. Insects were fed a standard production diet (Cohen, 2000). Last-instar male rather than female nymphs were used for library construction with the expectation of maximizing identification of feeding and digestive sequences and minimizing identification of egg development and maturation sequences. Male nymphs were collected fresh and total RNA was extracted using the Multi-enzymatic liquefaction of tissue (MELT™) total nucleic acid isolation system (Ambion) kit, following instructions provided by the manufacturer. RNA was further processed using the Poly(A) Purist™ MAG magnetic poly(A) RNA purification kit (Ambion), according to instructions provided. A cDNA library was constructed using the SuperScript™ plasmid system with Gateway® technology for cDNA synthesis and cloning (Invitrogen), according to instructions provided. Fractionated cDNA was ligated into the pSPORT1 vector and then introduced into chemically competent cells. Colonies were manually picked and transferred to 96-well plates for sequencing. Sequencing was performed by the USDA ARS MidSouth Genomics Core Facility. Sequences were analyzed by comparison with the tBLASTx algorithm (Altschul et al., 1997). RESULTS AND DISCUSSION We found many previously unknown genes for an increasingly important pest. Two hundred and seventy-two unique ESTs were generated, 48% of which could be identified as highly similar (expected value less than 1 × 10-10). The average sequence length was 768 bases, with only 11 ESTs having a length of less than 300 bases. Sequences less than 200 bases were not deposited as ESTs. Twenty-four of the sequences represented contigs, with the remainder as singlets. Accession numbers assigned to ESTs were DY470827 through DY470922, DY473180 through DY473230, and DY524480 through DY524596. Twelve sequences that appeared to represent complete or nearly complete genes, including actins, tubulins, cathepsins, pectinases, and a hexamerin, were submitted as standard GenBank submissions (accession numbers DQ386914, DQ450899, DQ471300, DQ471301, DQ399525-DQ399527, and DQ474245-DQ474249). Sequences that could be identified to an expected value of less than 1 × 10-25 were categorized into six groups (Table 1).
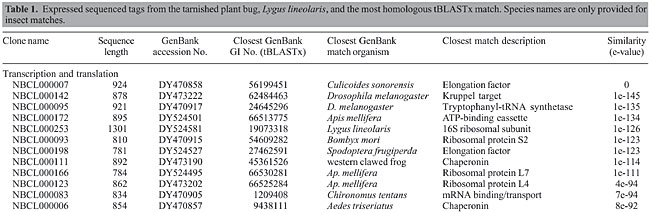        Sequences that corresponded to L. lineolaris genes that were already identified and deposited in GenBank were found, including 16S ribosomal sequence and a digestive enzyme trypsin (Zhu et al., 2003). We identified several gene sequences that may serve as endogenous controls for expression studies. ESTs from the transcription and translation group include ribosomal and mitochondrial sequences that have been useful for phylogenetic and population studies of arthropods (Murrell et al., 2001). Sequences that may be associated with feeding and digestion are of particular interest, especially the pectin-degrading enzymes identified as the primary cause of plant damage by this insect (Strong and Kruitwagen, 1968; Shackel et al., 2005). Additional putative digestive enzymes were identified by cross referencing the GenBank homologue with protein databases, as described in a study of another phygophagous insect (Pedra et al., 2003). Gene sequences previously identified from L. lineolaris include ribosomal sequences (Whiting et al., 1997), cytochrome oxidase (Tilmon et al., 2003), and cytochrome P450 monooxygenase sequences (Zhu and Snodgrass, 2003), odorant binding sequences (Vogt et al., 1999), esterases (Zhu et al., 2004), and trypsins (Zeng et al., 2002; Zhu et al., 2003). These previously submitted sequences were not duplicated in our submissions. Our purpose was to provide a broader window into the genetic composition of this important pest insect. This is the first EST library publicly deposited for this pest, and it includes genes associated with the damage-producing activities of the insect, namely feeding. There were 272 different ESTs generated, of which 260 were deposited in the NCBI dbEST repository. Twelve genes were deposited into GenBank as standard Core Nucleotide submissions. The mitochondrial EST sequences identified may be used to construct a whole mitochondrial genome sequence for this insect. Many sequences will be used in future research as endogenous controls, and to characterize functional genes for this pest and related species. ACKNOWLEDGMENTS I thank Fannie Williams and Catherine Smith for maintaining the Lygus lineolaris colony and for technical assistance. Insects for colony establishment were kindly provided by Eric Villavaso and Brenda Woods. Sequencing and batch bioinformatics processing were kindly provided by Brian Scheffler, Mary Duke, Linda Ballard, and Xiaofen Liu. I thank Douglas Streett, Eric Riddick, and Nathan Schiff for their comments on an earlier version of this manuscript. The United States Government has the right to retain a non-exclusive, royalty-free license in and to any copyright of this article. This article reports the results of research only. Mention of a commercial or proprietary product does not constitute an endorsement of the product by the United States Department of Agriculture. REFERENCES Altschul SF, Madden TL, Schaffer AA, Zhang J, et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. Cohen A (2000). New oligidic production diet for Lygus hesperus Knight and L. lineolaris (Palisot de Beauvois). J. Entomol. Sci. 35: 301-310. Murrell A, Campbell NJ and Barker SC (2001). A total-evidence phylogeny of ticks provides insights into the evolution of life cycles and biogeography. Mol. Phylogenet. Evol. 21: 244-258. Pedra JH, Brandt A, Westerman R, Lobo N, et al. (2003). Transcriptome analysis of the cowpea weevil bruchid: identification of putative proteinases and alpha-amylases associated with food breakdown. Insect Mol. Biol. 12: 405-412. Shackel KA, de la Paz Celorio-Mancera, Ahmadi H, Greve LC, et al. (2005). Micro-injection of Lygus salivary gland proteins to simulate feeding damage in alfalfa and cotton flowers. Arch. Insect Biochem. Physiol. 58: 69-83. Strong FE and Kruitwagen EC (1968). Polygalacturonase in the salivary apparatus of Lygus hesperus (Hemiptera). J. Insect Physiol. 14: 1113-1119. Tilmon KJ, Danforth BN, Day WH and Hoffman MP (2003). Determining parasitoid species composition in a host population: a molecular approach. Ann. Entomol. Soc. Am. 93: 640-647. Vogt RG, Callahan FE, Rogers ME and Dickens JC (1999). Odorant binding protein diversity and distribution among the insect orders, as indicated by LAP, an OBP-related protein of the true bug Lygus lineolaris (Hemiptera, Heteroptera). Chem. Senses 24: 481-495. Wheeler AG (2001). Biology of the plant bugs (Hemiptera: Miridae): pests, predators, opportunists. Comstock Publishing Associates, Ithaca. Whiting MF, Carpenter JC, Wheeler QD and Wheeler WC (1997). The Strepsiptera problem: phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology. Syst. Biol. 46: 1-68. Williams MR (2005). National cotton council: cotton insect losses estimates. http://www.msstate.edu/Entomology/Cotton.html. Accessed July 3, 2006. Zeng F, Zhu Y and Cohen A (2002). Partial characterization of trypsin-like protease and molecular cloning of a trypsin-like precursor cDNA in salivary glands of Lygus lineolaris. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 131: 453-463. Zhu YC and Snodgrass GL (2003). Cytochrome P450 CYP6X1 cDNAs and mRNA expression levels in three strains of the tarnished plant bug Lygus lineolaris (Heteroptera: Miridae) having different susceptibilities to pyrethroid insecticide. Insect Mol. Biol. 12: 39-49. Zhu YC, Zeng F and Oppert B (2003). Molecular cloning of trypsin-like cDNAs and comparison of proteinase activities in the salivary glands and gut of the tarnished plant bug Lygus lineolaris (Heteroptera: Miridae). Insect Biochem. Mol. Biol. 33: 889-899. Zhu YC, Snodgrass GL and Chen MS (2004). Enhanced esterase gene expression and activity in a malathion-resistant strain of the tarnished plant bug, Lygus lineolaris. Insect Biochem. Mol. Biol. 34: 1175-1186. |
|