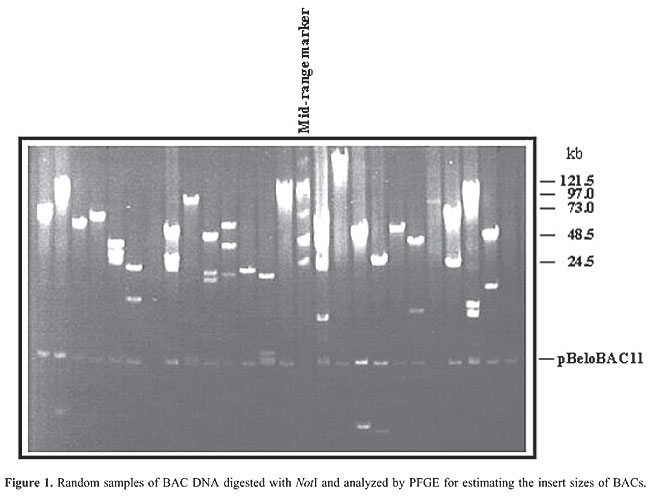
ABSTRACT. Corynebacterium pseudotuberculosis is a gram-positive bacterium that causes caseous lymphadenitis in sheep and goats. However, despite the economic losses caused by caseous lymphadenitis, there is little information about the molecular mechanisms of pathogenesis of this bacterium. Genomic libraries constructed in bacterial artificial chromosome (BAC) vectors have become the method of choice for clone development in high-throughput genomic-sequencing projects. Large-insert DNA libraries are useful for isolation and characterization of important genomic regions and genes. In order to identify targets that might be useful for genome sequencing, we constructed a C. pseudotuberculosis BAC library in the vector pBeloBAC11. This library contains about 18,000 BAC clones, with inserts ranging in size from 25 to 120 kb, theoretically representing a 390-fold coverage of the C. pseudotuberculosis genome (estimated to be 2.5-3.1 Mb). Many genomic survey sequences (GSSs) with homology to C. diphtheriae, C. glutamicum, C. efficiens, and C. jeikeium proteins were observed within a sample of 215 sequenced clones, confirming their close phylogenetic relationship. Computer analyses of GSSs did not detect chimeric, deleted, or rearranged BAC clones, showing that this library has low redundancy. This GSSs collection is now available for further genetic and physical analysis of the C. pseudotuberculosis genome. The GSS strategy that we used to develop our library proved to be efficient for the identification of genes and will be an important tool for mapping, assembly, comparative, and functional genomic studies in a C. pseudotuberculosis genome sequencing project that will begin this year. Key words: Bacterial artificial chromosome library, Corynebacterium pseudotuberculosis, Genomic survey sequence INTRODUCTION Corynebacterium pseudotuberculosis, a gram-positive intracellular pathogen, is the causative agent of caseous lymphadenitis in sheep and goats. The widespread occurrence and the economic importance of this disease have prompted investigation of its pathogenesis. However, the genetic basis of C. pseudotuberculosis virulence is still unknown (Dorella et al., 2006). The massive accumulation of prokaryotic DNA sequences generated by microbial genome projects allows us to rapidly investigate biological features and identify new molecular targets that might be useful for the development of immunological or chemotherapeutic reagents against any organism of choice (Celestino et al., 2004). Construction and characterization of a C. pseudotuberculosis library will facilitate further genetic studies of this organism, including the sequencing of its entire genome. Large insert libraries in bacterial artificial chromosome (BAC) vectors are particularly important, not only for genome sequencing projects, but also for comparative and functional genomic studies (Battistoni et al., 2005). The physical maps constructed with BACs are still widely used in genome research (Zhang and Wing, 1997). The BAC system is based on the well-studied Escherichia coli F factor. Replication of the F factor in E. coli is strictly controlled, since the F plasmid is maintained in low copy number (one or two per cell), thus reducing the potential for recombination between DNA fragments carried by the plasmid. F factors carrying inserted bacterial DNA are capable of maintaining fragments as large as 1 Mb, suggesting that the F factor is suitable for cloning large DNA fragments (Shizuya et al., 1992; Tillet and Neilan, 1998). The choice of the BAC cloning system allows us not only to cover the entire genome with a relatively small number of clones, but also to reduce the potential for recombination between DNA fragments, and more importantly, to avoid lethal overexpression of cloned bacterial genes (Amemiya et al., 1999). The BAC system allows us to clone large DNA molecules from a variety of complex genomic sources into bacteria, in which the DNA is stable, easy to manipulate, and is from a single foreign DNA source. The genetic stability of cloned DNA may be one of the most important aspects of this system (Shizuya et al., 1992; Zhang et al., 1996; Simon, 1997). BAC libraries of genomic DNA from numerous eukaryotic species have been constructed, and this has become the preferred approach for sequencing projects (Shizuya et al., 1992; Zhang et al., 1996; Zhang and Wing, 1997; Rogel-Gaillard et al., 1999). Furthermore, genomic libraries are a powerful resource for genetic studies of bacteria; there are various examples of successful use of this technology (Philipp et al., 1996; Brosch et al., 1998; Capela et al., 1999; Gordon et al., 1999; Brodin et al., 2002; Tauch et al., 2002, 2005; Feng et al., 2005; Walter et al., 2005). We describe here the construction of a C. pseudotuberculosis BAC library in pBeloBAC11 (Shizuya et al., 1992) and the beginning of the characterization of this library through a BAC-end sequencing strategy, namely genomic survey sequencing (GSS) (Kelley et al., 1998; Zhao, 2000; Ripoll et al., 2000). MATERIAL AND METHODS Bacterial strains, growth conditions and plasmid We used the bacterial strain C. pseudotuberculosis 1002, a wild strain provided by Dr. Roberto Meyer from the “Corynebacterium pseudotuberculosis Culture Collection”, Laboratory of Immunology (Federal University of Bahia); identification was confirmed by biochemical tests (API CORYNE, Biomerieux, Marcy l’Etoile, France). The E. coli strain used was E. coli DH10B (F– mcrA D (mrr-hdsRMS-mcrBC) f80d LacZ DM15 DLac X74 endA1 recA1 deoR D (ara,leu)7697 araD139 galU galK nupG rpsL l–; Grant et al., 1990). The plasmid pBeloBAC11 was as previously described (Shizuya et al., 1992). Corynebacterium pseudotuberculosis 1002 was aerobically grown in 2X YT (Difco Laboratories, Detroit, MI, USA) medium. E. coli was aerobically grown at 37°C in Luria-Bertani medium (LB, Difco Laboratories). When necessary, the medium was supplemented with chloramphenicol (12.5 µg/mL). BAC vector preparation Plasmid pBeloBAC11 preparation was carried out as described by Woo et al. (1994). Preparation of source chromosomal DNA High-molecular weight C. pseudotuberculosis DNA used in library construction was prepared on agarose plugs according to Azevedo et al. (1993). Plugs were stored in 50 mM EDTA at 4°C and washed three times in 0.1% Triton X-100 buffer prior to use. Partial digestion and size selection were carried out as previously described (Brosch et al., 1998). Cloning of C. pseudotuberculosis DNA into pBeloBAC11 Agarose containing the selected fragments was melted at 70°C for 5 min and digested with GELase (Epicentre Technologies, Madison, WI, USA), using 1 U per 100 µL of gel slice. Then, a C. pseudotuberculosis library was constructed by ligation of partially digested HindIII fragments (25-120 kb) into a previously prepared pBeloBAC11 vector, in a 1:10 molar ratio using 5 U T4 Ligase (Invitrogen Ltd., CA, USA), at 16°C for 18 h. Ligation mixtures were heated to 65°C for 15 min and then drop-dialyzed against Tris-EDTA, using VS 0.025 mM membranes (Millipore, Bedford, MA, USA). Transformation of E. coli DH10B was carried out by electroporation. Competent cells were prepared; electroporation and selection of recombinants were performed according to Brosch et al. (1998). Each selected clone (white colony) was inoculated manually into 96-well plates, containing 100 µL LB plus chloramphenicol (12.5 µg/mL) and incubated at 37°C overnight. After incubation, 100 µL of 80% glycerol was added to each well; the plates were sealed with polyester film and stored at -70°C. Sizing of BAC DNA inserts BAC DNA from recombinant clones was isolated by the alkaline lysis method, as previously described (Birnboin and Doly, 1979), with minor modifications. To estimate the insert size, BAC DNA was digested with NotI and analyzed by PFGE using a 1% Agarose SeqPlaque gel in 1X TAE buffer and 0.5 µg/mL ethidium bromide at 10°C, with a ramp time from 4 s at 6.25 V/cm for 15 h on an LKB-Pharmacia CHEF apparatus. The MidRange II PFGE marker (New England Biolabs) was used as a size standard. GSS generation through BAC-end sequencing Sequencing reactions were performed using the Taq DyeDeoxy Terminator cycle sequencing kit in an ABI PrismTM 377 DNA Sequencer (Applied Biosystems, Foster City, CA, USA). Sequencing primers were M13 (-40, forward) 5’ GTT TTC CCA GTC ACG AC 3’ and M13 (reverse) 5’ CAG GAA ACA GCT ATG AC 3’. One microgram of DNA and 5 µM of the primers were added to 14-µL reaction mixtures containing 5 µL of dye terminator premix. Sequencing reactions were cycled according to the following program: 5 min at 95°C and 40 cycles of 20 s at 95°C, 15 s at 55°C and 1 min at 60°C. Reaction products were purified as follows: 120 µL 75% isopropanol (v/v) was added to samples, which were incubated in the dark for 20 min at room temperature. The samples were then centrifuged at 14 rpm for 20 min and the supernatants were discarded; 100 µL 70% ethanol (v/v) was added and the resulting mixture was centrifuged at 14 rpm for 10 min. The supernatants were discarded and the DNA was resuspended in 10 µL deionized formamide. Bioinformatics Similarity searches of each BAC-end sequence were made with the “Basic Local Alignment Search Tool” software (http://www.ncbi.nlm.nih.gov/blast) service of the National Center for Biotechnology Information (NCBI). GSSs were classified according to BLAStx analysis. Only those GSSs with a positive match [best BLAST hit with an expected value (E) of <10-10 and score >42] for a protein in the non-redundant database were considered. Nucleotide sequence accession numbers The nucleotide sequences that were identified were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/Genbank/) under accession numbers BH740422 to BH740650. RESULTS AND DISCUSSION A genomic library with approximately 18,000 clones was constructed as described in Material and Methods, with inserts with an average size of 25 to 120 kb (Figure 1). A random sample of 215 clones was successfully end sequenced and deposited in the GenBank databases.
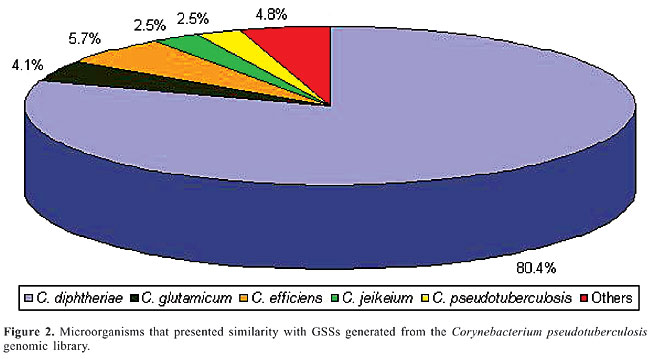
After deleting vector sequences and unreliable data, an average of 359 bases per clone was obtained and used for database searches. These data theoretically represent a 390-fold coverage of the C. pseudotuberculosis genome (estimated to be 2.5-3.1 Mb; Redenbach et al., 2000). The redundancy of the library was estimated to be 13% through sequence alignment using the CAP3 software (http://pbil.univ-lyon1.fr/cap3.php). A high-quality genomic library must display a value lower than 20% redundant sequences, clones without inserts or clones with contamination. Sequence similarities identified by BLASTx were considered statistically significant (E) at <10-10 and score >42. Among the 229 GSSs obtained, 123 (53.7%) had a significant match with proteins of the GenBank database. Table 1 shows the best BLASTx hits obtained in the similarity searches; the organisms were ordered according to the degree of conservation. A significant number of valid GSSs showed similarity with C. diphtheriae, C. glutamicum, C. efficiens, and C. jeikeium proteins, confirming their close phylogenetic relationship (Figure 2). Similarity with proteins from other groups was also observed. Three GSSs presented similarity with C. pseudotuberculosis sequences (1 for 16S rRNA and 2 for RpoB), validating this method for C. pseudotuberculosis.       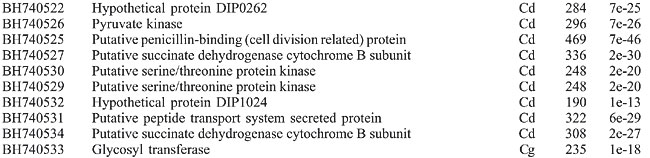     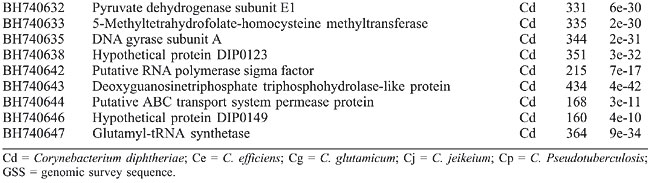
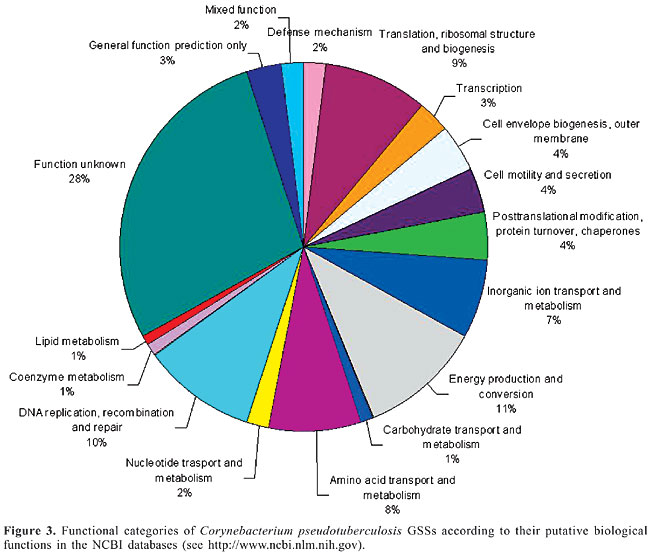
For an insight into the functional diversity of our random sequences, we compared our GSSs to the sequences present in the database and derived the functional classification associated with each protein. We found 123 GSSs that could be classified into 17 broad functional categories (Figure 3). Some GSSs were classified into more than one category and thus were included in the “mixed function” group. The largest fraction (28%) was placed in the function unknown group, which includes hypothetical proteins. Other important categories included sequences related to energy production and conversion (11%), DNA replication, recombination and repair (10%) and translation, ribosomal structure and biogenesis (9%).
CONCLUSIONS Our libraries proved to be an important tool for mapping and genome assembly and could be used in a genome-sequencing project. The construction of this genomic library was part of the preparatory work for the Minas Gerais C. pseudotuberculosis (strain 1002) chromosome sequencing project, which will be initiated this year (2006). ACKNOWLEDGMENTS Research supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil), FINEP (Financiadora de Estudos e Projetos - 01.04.760.00), and FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais, Brazil). REFERENCES Amemiya CT, Zhong TP, Silverman GA and Fishman MC (1999). The zebrafish, genomics and genetics. In: Methods in cell biology (Detrich W, Westerfield M and Zon L, eds.). Academic Press, New York, 235-258. Azevedo V, Alvarez E, Zumstein E, Damiani G, et al. (1993). An ordered collection of Bacillus subtilis DNA segments cloned in yeast artificial chromosomes. Proc. Natl. Acad. Sci. USA 90: 6047-6051. Battistoni F, Bartels D, Kaiser O, Marie Reamon-Buettner S, et al. (2005). Physical map of the Azoarcus sp. strain BH72 genome based on a bacterial artificial chromosome library as a platform for genome sequencing and functional analysis. FEMS Microbiol. Lett. 249: 233-240. Birnboim HC and Doly J (1979). A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7: 1513-1523. Brodin P, Eiglmeier K, Marmiesse M, Billault A, et al. (2002). Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect. Immun. 70: 5568-5578. Brosch R, Gordon SV, Billault A, Garnier T, et al. (1998). Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect. Immun. 66: 2221-2229. Capela D, Barloy-Hubler F, Gatius MT, Gouzy J, et al. (1999). A high-density physical map of Sinorhizobium meliloti 1021 chromosome derived from bacterial artificial chromosome library. Proc. Natl. Acad. Sci. USA 96: 9357-9362. Celestino PBS, Carvalho LR, Freitas LM and Dorella FA (2004). Update of microbial genome programs for bacteria and archaea. Genet. Mol. Res. 3: 421-431. Dorella FA, Pacheco LG, Oliveira SC, Miyoshi A, et al. (2006). Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet. Res. 37: 201-218. Feng Z, Qi J, Tsuge T, Oba Y, et al. (2005). Construction of a bacterial artificial chromosome library for a myxobacterium of the genus Cystobacter and characterization of an antibiotic biosynthetic gene cluster. Biosci. Biotechnol. Biochem. 69: 1372-1380. Gordon SV, Brosch R, Billault A, Garnier T, et al. (1999). Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32: 643-655. Grant SG, Jessee J, Bloom FR and Hanahan D (1990). Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87: 4645-4649. Kelley JM, Field CE, Craven MB, Bocskai D, et al. (1999). High throughput direct end sequencing of BAC clones. Nucleic Acids Res. 27: 1539-1546. Philipp WJ, Nair S, Guglielmi G, Lagranderie M, et al. (1996). Physical mapping of Mycobacterium bovis BCG Pasteur reveals differences from the genome map of Mycobacterium tuberculosis H37Rv and from M. bovis. Microbiology 142 (Pt 11): 3135-3145. Redenbach M, Scheel J and Schmidt U (2000). Chromosome topology and genome size of selected actinomycetes species. Antonie Leeuwenhoek 78: 227-235. Ripoll PJ, O’Sullivan DM, Edwards KJ and Rodgers M (2000). Technique for cloning and sequencing the ends of bacterial artificial chromosome inserts. Biotechniques 29: 271-276. Rogel-Gaillard C, Bourgeaux N, Billault A, Vaiman M, et al. (1999). Construction of a swine BAC library: application to the characterization and mapping of porcine type C endoviral elements. Cytogenet. Cell Genet. 85: 205-211. Shizuya H, Birren B, Kim UJ, Mancino V, et al. (1992). Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. USA 89: 8794-8797. Simon MI (1997). Dysfunctional genomics: BACs to the rescue. Nat. Biotechnol. 15: 839. Tauch A, Homann I, Mormann S, Ruberg S, et al. (2002). Strategy to sequence the genome of Corynebacterium glutamicum ATCC 13032: use of a cosmid and a bacterial artificial chromosome library. J. Biotechnol. 95: 25-38. Tauch A, Kaiser O, Hain T, Goesmann A, et al. (2005). Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J. Bacteriol. 187: 4671-4682. Tillett D and Neilan BA (1998). Small-scale preparation of the single-copy bacterial artificial chromosome vector pBeloBAC11. Biotechniques 24: 568-572. Walter J, Mangold M and Tannock GW (2005). Construction, analysis, and beta-glucanase screening of a bacterial artificial chromosome library from the large-bowel microbiota of mice. Appl. Environ. Microbiol. 71: 2347-2354. Woo SS, Jiang J, Gill BS, Paterson AH, et al. (1994). Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 22: 4922-4931. Zhang HB and Wing RA (1997). Physical mapping of the rice genome with BACs. Plant Mol. Biol. 35: 115-127. Zhang HB, Woo SS and Wing RA (1996). BAC, YAC and cosmid library construction. In: Plant gene isolation: principles and practice (Foster GD and Twell D, eds.). John Wiley & Sons Ltd., Chichester, West Sussex, England, 75-99. Zhao S (2000). Human BAC ends. Nucleic Acids Res. 28: 129-132. |
|