ABSTRACT. All organisms that have been studied until now have been found to have differential distribution of simple sequence repeats (SSRs), with more SSRs in intergenic than in coding sequences. SSR distribution was investigated in Archaea genomes where complete chromosome sequences of 19 Archaea were analyzed with the program SPUTNIK to find di- to penta-nucleotide repeats. The number of repeats was determined for the complete chromosome sequences and for the coding and non-coding sequences. Different from what has been found for other groups of organisms, there is an abundance of SSRs in coding regions of the genome of some Archaea. Dinucleotide repeats were rare and CG repeats were found in only two Archaea. In general, trinucleotide repeats are the most abundant SSR motifs; however, pentanucleotide repeats are abundant in some Archaea. Some of the tetranucleotide and pentanucleotide repeat motifs are organism specific. In general, repeats are short and CG-rich repeats are present in Archaea having a CG-rich genome. Among the 19 Archaea, SSR density was not correlated with genome size or with optimum growth temperature. Pentanucleotide density had an inverse correlation with the CG content of the genome. Key words: CG content, Microsatellites, SSRs, Hyperthermophiles, Thermophiles, Optimum growth temperature INTRODUCTION Microsatellites, also named simple sequence repeats (SSRs), are widespread throughout eukaryote, prokaryote and virus genomes. SSR frequency and distribution are species and motif specific (Karlin et al., 1997; Bachtrog et al., 1999, 2000; Butcher et al., 2000; Crollius et al., 2000; Metzgar et al., 2000; Toth et al., 2000; Gentles and Karlin, 2001; Morgante et al., 2002). Mechanisms for SSR genesis include transpositions, insertions, horizontal gene transfer, recombination and repair, in addition to slippage during replication (Primmer and Ellegren, 1998; Hancock and Santibanez-Koref, 1998; Hartenstine et al., 2000; Chambers and MacAvoy, 2000; Schlotterer, 2000; Jakupciak and Wells, 2000; Zhu et al., 2000; Alba et al., 1999a,b, 2001). However, repeat expansions may be orientation or strand specific and may be independent of the efficiency of the repair system (Morel et al., 1998; Cleary et al., 2002). Since there appears to be similarity in these findings in prokaryotes and eukaryotes, the method of repeat generation apparently has not changed with time (Achaz et al., 2002). Comparative SSR distribution in the Archaea has not been studied in detail until now; studies have been restricted to repeats other than SSRs (Cox and Mirkin, 1997; Karlin et al., 1997; Smith et al., 1997; Fitz-Gibbon et al., 2002). Study of SSR profile in the Archaea is appealing, because they are enigmatic organisms occupying diverse habitats, including extreme environments. Although they have similarities with eubacteria and eukaryotes, the presence of unique features in the Archaea has maintained the debate over whether they are intermediates or ancestors of both pro- and eukaryotes (Woese et al., 1990; Doolittle, 1995; Zlatanova, 1997; Makarova and Koonin, 2003). The genome sequence data that are now available make it possible to conduct a comparative analysis of SSRs. SSR density and motif types in complete sequences of the main chromosome of 19 Archaea were examined and compared. MATERIAL AND METHODS The main chromosome sequences of 19 Archaea (Table 1) were downloaded from the NCBI database (www.ncbi.nlm.nih.gov). Complete chromosome sequences were used for analysis, without removing r-RNA or t-RNA sequences. Plasmid sequences were not included in the analysis. Tandem repeat tracts (dinucleotide to pentanucleotide) were obtained by analyzing complete sequences with the program “SPUTNIK” (C. Abajian, University of Washington, http://www.abajian.com/sputnik) with some modifications to accommodate the analysis of larger sequences, which was not possible with the original version. More recently, the site link to SPUTNIK changed to http://espressosoftware.com/pages/sputnik.jsp, after this study was completed.
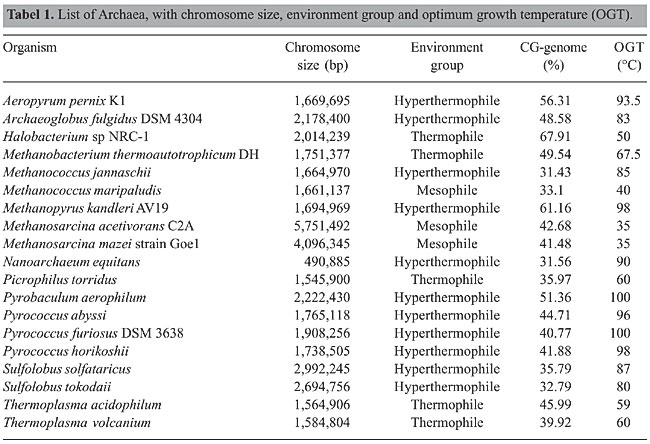
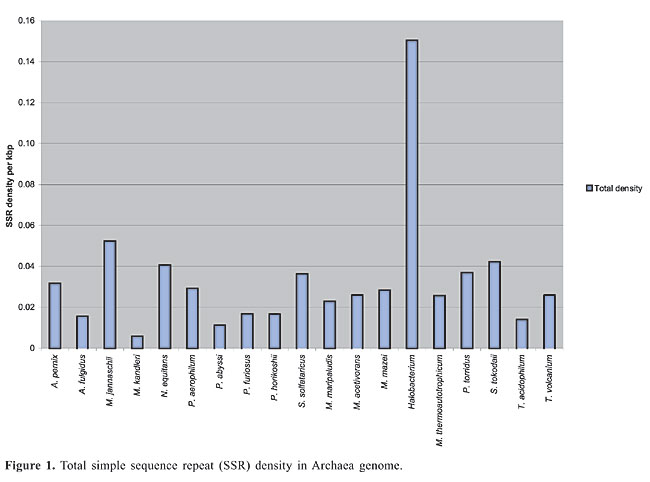
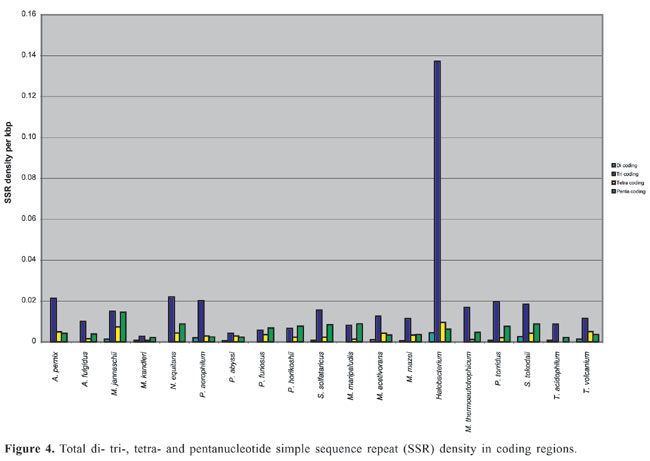
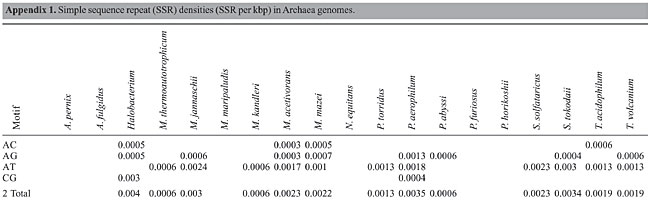
The density of SSRs was calculated by dividing the number of SSRs by the total sequence length of the main chromosome. Density in the coding (m-RNA, t-RNA and r-RNA) and non-coding regions was calculated by dividing the number of SSRs by the total coding sequence and intergenic sequence lengths, respectively. Total coding sequences were taken from protein and RNA tables of the NCBI database for each organism. Overlapping sequences in coding regions were removed for calculating the total coding sequences. The term “Horizon” applies to SSRs found partially in the intergenic region and partially in the coding region and because of uncertainty whether they lie in untranscribed or untranslated regions (UTR). These SSRs were considered in the calculation of total SSR density but not in coding and intergenic regions. I only included simple sequence repeats, classified as microsatellites. The search was also limited to di-, tri-, tetra- and pentanucleotide repeats. All possible combinations of repeats were grouped together. For example, (AC)n, (CA)n, (GT)n, and (TG)n in the case of dinucleotide repeats and (CCG)n, (GCC)n, (CGG)n, and (GGC)n in the case of trinucleotide repeats were grouped together, even though these groupings may result in losing information about amino acid preferences in the coding region. To determine repeat length, the total number of base pairs of an SSR was counted. For example, for the dinucleotide CG repeat “CGCGCGCGCGCG”, the length was calculated as 12 bp. The average total length thus obtained for each motif and repeat class was used for statistical analysis. Pearson correlation analysis (two-tailed) using Microsoft Excel functions was used to examine the correlation of SSR density with chromosome size, CG content of the genome, optimum growth temperature (OGT), repeat length, and repeat CG richness. Archaea OGTs were obtained from the Prokaryotic Growth Temperature database “http://pgtdb.csie.ncu.edu.tw/” (Table 1). Archaeoglobus fulgidus, which was classified as a thermophile in the database, was grouped as a hyperthermophile in the present study because the OGT lies in this range. RESULTS Density of SSR and most common motifs in the genome The complete main chromosome sequences (henceforth referred to as genomes) of 19 Archaea revealed diversity in the distribution of SSRs, with densities ranging from 0.1504/kbp in Halobacterium to 0.0059/kbp in M. kandleri (Appendix 1 and Figure 1). Among SSRs, trinucleotide repeats (ACG, CAG, CGC, CCG, and AGC in particular) have the highest density (Appendix 1). SSRs are present in coding and non-coding regions of all Archaea. Archaeoglobus fulgidus, M. kandleri, P. abyssi, P. horikoshii, and Halobacterium have higher densities of repeats in coding sequences than in non-coding regions (Figure 2).
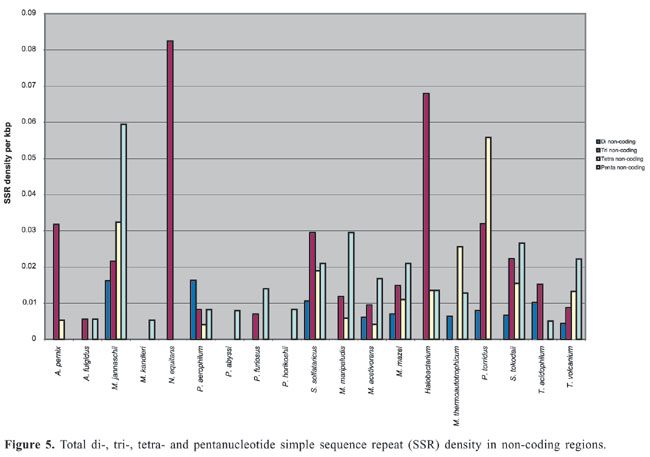
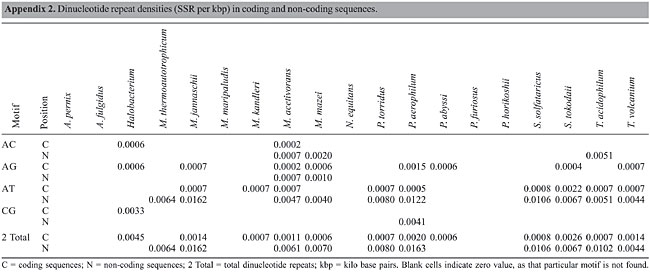
Dinucleotide repeats were found in most of the Archaea genomes, except in A. pernix, A. fulgidus, M. maripaludis, N. equitans, P. furiosus, and P. horikoshii and in the coding regions of M. thermoautotrophicum (Appendixes 1 and 2 and Figures 3 and 4). Dinucleotide repeats were found to be abundant in non-coding regions as compared to coding regions (Appendix 2 and Figures 4 and 5). AT repeats were most common, followed by AG repeats in the total genome as well as in coding and non-coding regions. AC repeats were not common and CG repeats were rare.
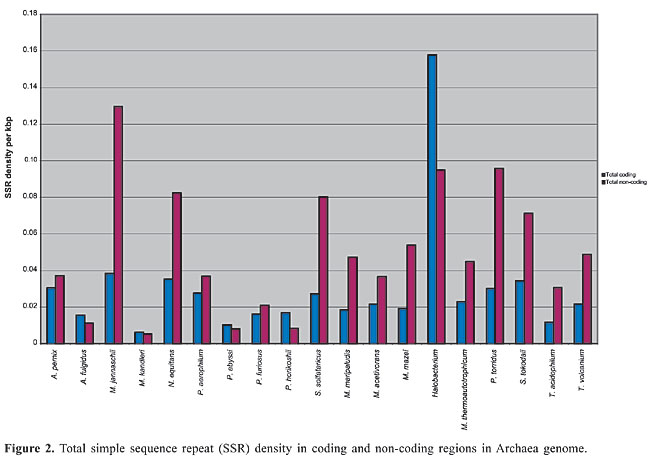
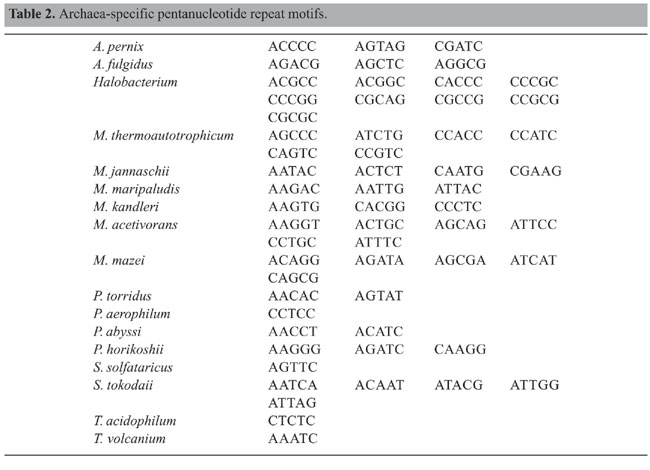
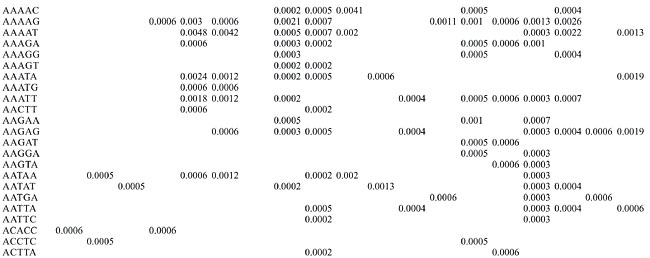
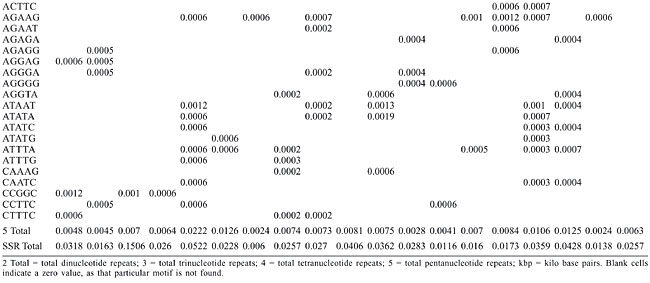
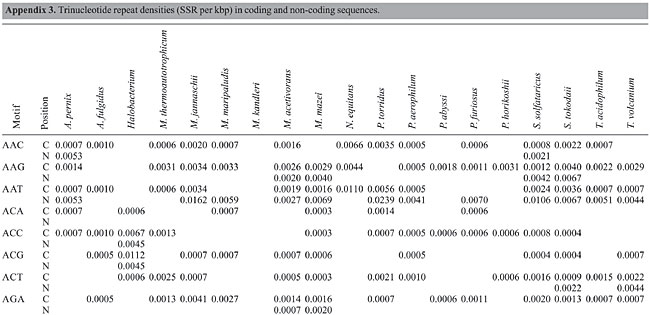
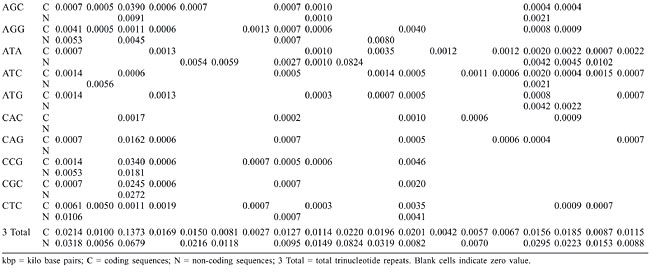
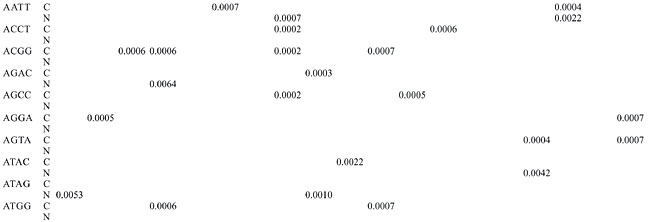
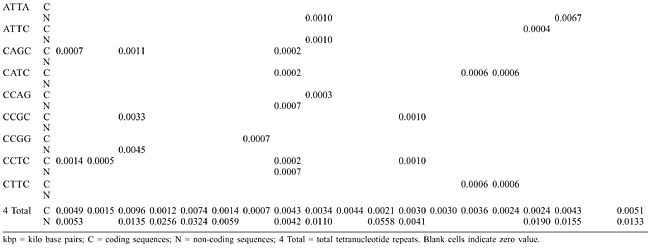
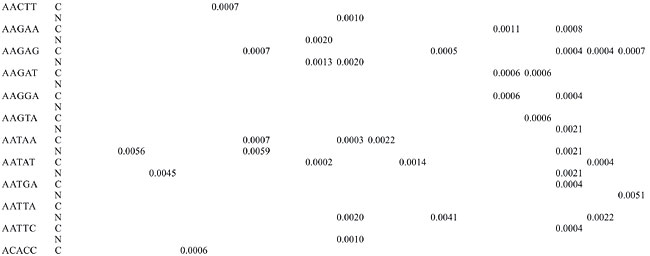
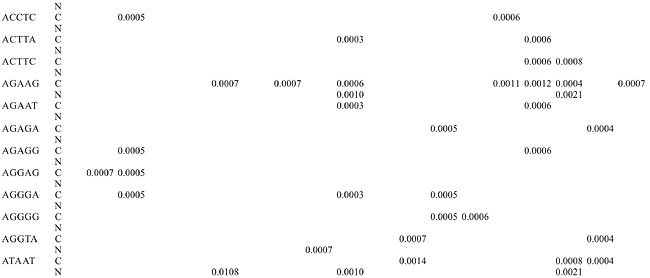
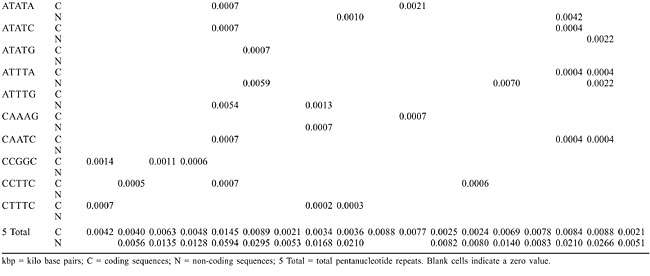
Trinucleotide repeats in the total genome outnumbered all other repeats in almost all of the Archaea genomes, except M. jannaschii, P. abyssi, P. furiosus, P. horikoshii, and M. maripaludis, where pentanucleotide repeats were more frequent than trinucleotide repeats (Figure 3). Most of the Archaea genomes have AAT and AAG mofits. Coding regions of most Archaea have AAG mofit and non-coding regions have AAT mofits (Appendixes 1 and 3). In coding regions, AAT was the most abundant repeat in N. equitans, P. torridus and S. solfataricus. AAG was the most abundant repeat in M. thermoautotrophicum, M. maripaludis, M. acetivorans, M. mazei, P. abyssi, P. horikoshii, S. tokodaii, T. acidophilum, and T. volcanium. AGC in Halobacterium, AGA in M. jannaschii, AGG in M. kandleri, CCG in P. aerophilum, and ATC in P. furiosus were the most abundant repeats. Trinucleotide repeats were not present in non-coding regions of M. thermoautotrophicum, M. kandleri, P. abyssi, and P. horikoshii (Appendix 3 and Figure 5). In non-coding regions, only the ATC repeat was present in A. fulgidus, ATA in N. equitans and AAT in P. furiosus intergenic regions. CTC was the most abundant repeat in A. pernix and P. aerophilum, ACT in T. volcanium and CGC in Halobacterium. AAT was the most abundant repeat in M. jannaschii, M. mazei, P. torridus, S. solfataricus, and S. tokodaii. ATA was the most abundant repeat in M. maripaludis, M. acetivorans and T. acidophilum. Tetranucleotide repeats were found in the total genome of all Archaea except T. acidophilum (Appendix 4 and Figure 3). Species-specific repeats include AACT, AATG, AGGC, and AATC in M. mazei, AAGC and ACAT in A. fulgidus, AGCG and AGGG in A. pernix, AGTC and CATG in M. acetivorans, ATTG, CCCG, CGAG, CGGC, and CACG in Halobacterium, and CAAG and CAGG in P. furiosus. These motifs and their respective densities are not shown in Appendixes 1 and 4. AAAT and AAGA were most common repeats, but some motifs that were found in the total genome were not present in coding regions (Appendixes 1 and 4). AACT, AATC, AATG, ACAT, ATAG, ATTA, ATTG, and CATG were absent in the coding sequences of all Archaea. Tetranucleotide repeats were not found in intergenic sequences of A. fulgidus, M. kandleri, N. equitans, P. abyssi, P. furiosus, and P. horikoshii (Appendix 4 and Figure 5). All nineteen Archaea were found to have pentanucleotide repeats in the total genome and coding regions, with diverse pattern prevalences (Appendixes 1 and 5 and Figures 3 and 4). Out of 84 repeat motifs, AAAAG was common but 58 were Archaea-specific (densities not shown in Table 2). The pentanucleotide density in coding regions of P. furiosus, P. horikoshii and M. maripaludis was higher than that of trinucleotide repeats (Figure 4). Non-coding regions of A. pernix, N. equitans and P. torridus did not have pentanucleotide repeats (Figure 5).
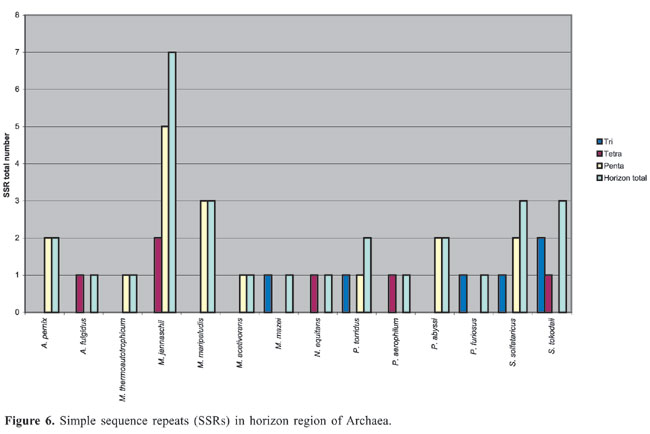
Horizon region SSRs were found partially in coding and partially in intergenic regions of 14 Archaea, among which the most frequent occurrence of SSRs (seven) was in M. jannaschii (Figure 6). Dinucleotides were not found in the overlapping regions. Halobacterium, M. kandleri, P. horikoshii, T. acidophilum, and T. volcanium did not have horizon SSRs.
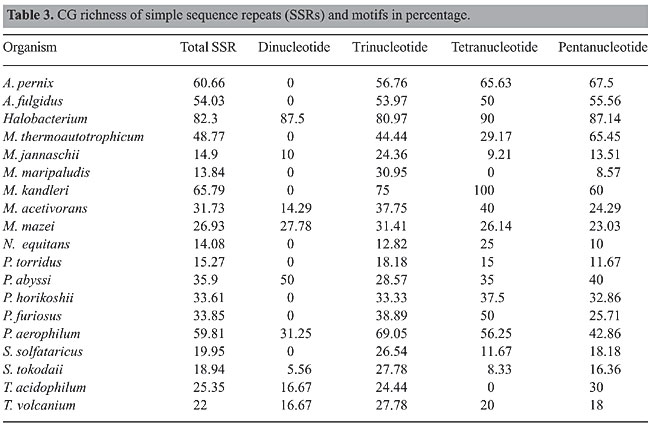
CG richness of SSRs CG richness of total SSRs, di-, tri- and penta-nucleotide repeats was highest in Halobacterium. Total SSRs and pentanucleotide repeats had lowest CG richness in M. maripaludis. CG richness of di- and tetranucleotide repeats was lowest in S. tokodaii, and N. equitans had the fewest trinucleotide repeats. Since M. kandleri had only one tetranucleotide repeat (CCGG), it shows 100% CG richness. However, Halobacterium had the second highest CG richness (Table 3).
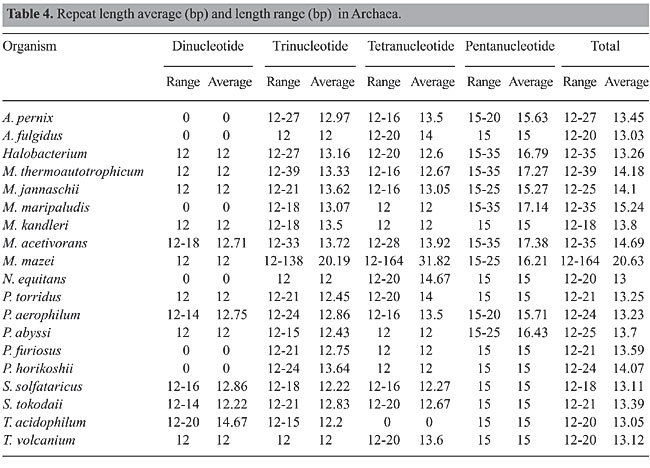
Repeat length Repeats were generally not long in Archaea, as the average minimum repeat length was 13 bases and the maximum was 20.63 bases (Table 4). Exceptions include M. mazei [164 (AATA), 138 (AAT), 60 (AAAT), 52 (AATG), and 51 (AAG and ATA) bases] and M. thermoautotrophicum (AGC 39 bp long), which have long repeats. The maximum dinucleotide repeat was 20 bases (AT in T. acidophilum). The minimum di-, tri- and tetra-nucleotide repeat lengths were 12 bases. The minimum pentanucleotide repeat length was 15 bases, but long repeats (35 bases) were found in Halobacterium (CGCAG), M. thermoautotrophicum (CAGTC), M. maripaludis (AAAAT), M. acetivorans (ATTTC), and M. acetivorans (AAATA).
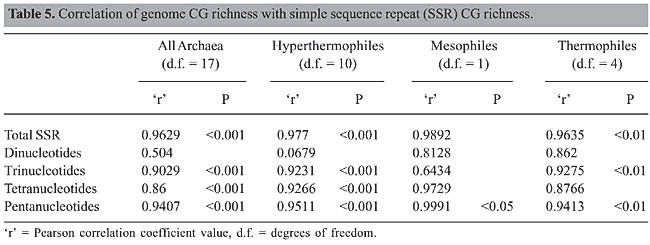
DISCUSSION The finding of 167 repeat motifs indicates that SSRs are not rare in Archaea genomes. These repeats show species-specific characteristic distributions, as has been reported for many other organisms (Toth et al., 2000; Rocha and Blanchard, 2002). Although repeat patterns have been previously studied in Archaea sequences (Morris et al., 1986; Smith et al., 1997; Rocha et al., 1999), it was not possible to compare those studies with the present one due to the fact that incomplete sequences were available at that time or there were differences in the sequence length analyzed and stringent length criteria for repeats in those studies. Dinucleotide repeats Abundant dinucleotide repeats were not found in the Archaea. This is similar to findings for other organisms (Karlin et al., 1997; Toth et al., 2000). AT repeats were found to be more frequent than CA/TG repeats in the Archaea, which is consistent with studies in P. aerophilum and Sulfolobus (Karlin et al., 1997), Aves genome and plants (Primmer et al., 1997; Toth et al., 2000; Morgante et al., 2002). However, it is contrary to findings in prokaryote and eukaryote sequences (Campbell et al., 1999) and in Arabidopsis thaliana (Morgante et al., 2002). Karlin et al. (1997) suggest that AT repeats have the ability to form less thermodynamically stable DNA duplexes, which are preferred sites for cleavage by RNAase in mRNA and could lead to inappropriate binding of regulatory proteins. Possibly, these could be the reasons for absence of TA repeats in some Archaea. Paradoxically, AT repeats could be responsible for increasing DNA flexibility and association with histone-like proteins, resulting in their playing important roles in gene regulation and chromatin folding (Okonogi et al., 2000). It remains to be investigated whether AT repeats play similar roles in Archaea. CG repeats are abundant only in Halobacterium, similar to what was found by Karlin et al. (1997). The absence of CG repeats in most Archaea cannot be due to methylation-driven mutations alone (Wang et al., 2004). This is because Drosophila, animal mitochondria and Neurospora lack methylase activity and yet TA are more common than CG repeats. Therefore, CG dinucleotide deficiency could be due to selective advantage, given structural constraints related to high stacking energy and chromatin packing (Karlin et al., 1997; Lerat et al., 2002) or to avoid blocking of transcription (Morris et al., 1986). Trinucleotide repeats An abundance of trinucleotide repeats was found, which could be due to mechanisms that suppress nontrimeric repeats (Metzgar et al., 2000). The most common repeats in Archaea are AAT, AAG and AGC, which corroborates the reported abundance of AAG repeats in plants and AGC in animals but contrary to the finding of rare AAT repeats in monocots (Varshney et al., 2002; Thiel et al., 2003). CAG and CCG repeats were found in seven Archaea, despite the fact that they are known to be highly unstable in many organisms (Moore et al., 1999; Jakupciak and Wells, 2000; Ireland et al., 2001; Hashem et al., 2002). The abundance of CAG repeats in Archaea-coding regions could be due to the influence of encoded amino acids (Alba et al., 1999a,b; Varshney et al., 2002; Thiel et al., 2003). CCG repeats are present in Archaea having >40% genome CG content. This could be due to the influence of the high CG content of their genomes (Morgante et al., 2002; Varshney et al., 2002; Thiel et al., 2003). CCG, ACA, CAC, and GGA repeats may be associated with protein folding/solubility, nucleosome proteins and stress response (Godde and Wolffe, 1996; Grayling et al., 1997; Mishima et al., 1997; Pereira et al., 1997; Reeve et al., 1997; Pereira and Reeve, 1998; Satyal et al., 2000; Sandman and Reeve, 2000, 2001). Green and Wang (1994) report that some trinucleotide repeats may be important for adding new coding regions, new functions to proteins, or for increasing the size of proteins. The latter may not be true in the case of thermophilic and hyperthermophilic Archaea, as the protein size is generally small in thermophiles (Chakravarty and Varadarajan, 2000; Hickey and Singer, 2004). The absence of some repeats, such as GGA, CCG, in some Archaea, in addition to the absence of ACA, CAC and CAG repeats in non-coding regions of all Archaea but presence in coding sequences, is an interesting feature. Tetranucleotide and pentanucleotide repeats Tetranucleotide repeats are not abundant in Archaea, but unlike in Escherichia coli (Rocha et al., 2002), they are not underrepresented. Exceptions are M. kandleri and T. acidophilum, which have one and no tetranucleotide repeats, respectively. Underrepresentation of CTAG, CATC and GTAC in Halobacterium and M. jannaschii agrees with the findings of Karlin et al. (1997). However, they reported a normal presence of these repeats in Sulfolobus, while none was found in the present study. Although tetra- and pentanucleotide repeat densities are more abundant in non-coding regions compared to coding regions, in some Archaea these repeats are absent in non-coding sequences. Pentanucleotide repeats in Archaea show characteristic distributions as diverse as in other organisms (Toth et al., 2000; Gur-Arie et al., 2000; Lim et al., 2004). Coding and non-coding regions Generally, repeats are more abundant in non-coding regions (Primmer et al., 1997; Primmer and Ellegren, 1998; Bachtrog et al., 1999; Elgar et al., 1999; Crollius et al., 2000; Gur-Arie et al., 2000; Dokholyan et al., 2000; Toth et al., 2000; Katti et al., 2001) to avoid the ill effects of repeat stability in coding regions (Schlotterer, 1998; Hancock and Santibanez-Koref, 1998; Harr et al., 1998, 2000; Ellegren, 2000; Chambers and MacAvoy, 2000; Dokholyan et al., 2000). Earlier studies suggested no functional roles of SSRs due to their presence in pseudogenes and intergenic sequences of P. aerophilum (Fitz-Gibbon et al., 2002). Abundance of repeats in coding regions of some Archaea is contrary to these findings, and it is evident that Bacillus subtilis (Rocha et al., 1999) is not exceptional in having abundance of SSRs in coding sequences. Trinucleotide repeats are abundant in coding regions in Archaea, like most organisms studied so far, because they may leave reading frames unperturbed (Karlin et al., 1997; Subramanian et al., 2003). However, an abundance of tetra- and pentanucleotide repeats was also found in coding sequences as well as their absence in non-coding sequences of many Archaea. Therefore, it is possible that SSRs are tolerated in coding regions because their variations affect not only gene expression but also adaptation to environmental factors in prokaryotes (see review by Li et al., 2004). It is known that the factors that affect preferential distribution of repeats in non-coding and coding regions work differentially in all organisms and the bias is marked in eukaryotes (Cox and Mirkin, 1997; Marcotte et al., 1999). Is it because the known or yet unknown functions of repeats in prokaryote (Archaea in particular)-coding regions are not important in higher organisms? It is possible that coding regions in eukaryotes do not have as many sites for insertions as in prokaryotes, or that there is little tolerance for integrations and hence fewer SSRs. This is because repeat genesis may be a result of insertional events due to transposons and virus (Ogura et al., 1994; Ramsay et al., 2000; Cardle et al., 2000; Lerat et al., 2002). However, why Archaea would tolerate SSRs in coding sequences remains uninvestigated, except for the fact the amino acid reiterations affect protein stability (Gromiha et al., 2002; de Farias and Bonato, 2002; Chakravarty and Varadarajan, 2002; Farias and Bonato, 2003), which would be an essential requirement for extremophilic Archaea. It would be a fruitful exercise to investigate common repeat motifs in prokaryotes and eukaryotes, and genes associated with SSRs to study the fate of specific patterns and coding regions that have retained or lost repeats in eukaryotes. In this light, genes related to DNA repair, recombination and adaptations to different types of stress (Rocha et al., 2002), genes associated with t-RNA, r-RNA, DNA repair and replication, gonads, silk glands and development in Bombyx mori (silkworm) have a high density of SSRs (Trivedi, 2003). Horizon region Dinucleotide repeats were found to be absent in the horizon region, different from the abundance of AG repeats in 5’UTR of plants and 3’UTR of catfish and the abundance of SSRs in UTR compared to coding sequences. Since SSRs are not preferred in the UTR in Archaea, they could not be important for gene regulation or silencing, protein adaptations, and transcription slippage, which results in long m-RNAs in other organisms (Stallings, 1995; Wren et al., 2000; Morgante et al., 2002). Genome size, SSR density and optimum growth temperature Nineteen Archaea, hyperthermophiles and thermophiles showed no correlation of OGT with either total SSR or motif density in genome, coding and non-coding regions. Similarly, total SSR and motif densities showed no correlation with genome size. However, a trend was seen in thermophiles, where pentanucleotide repeats correlated positively with genome size (‘r’ = 0.8374; P < 0.05); this should be examined when other thermophiles are considered for such analysis. The absence of correlation with genome size is consistent with earlier studies (Hancock, 2002; Lim et al., 2004). However, it is contrary to reports of a positive correlation between genome size and SSR density, the contribution of repeats to increased genome size and the C-value paradox in various organisms (Hancock, 1996a; Primmer et al., 1997; Achaz et al., 2002; Trivedi, 2004). Paradoxically, N. equitans has the smallest genome size but has the fourth-highest SSR density. If there was a reduction in genome size of this organism, it may not have been due to SSR elimination, even if repeats may be superfluous, as suggested for Mycoplasma genitalium (Hancock, 1996b). SSRs in Archaea of a given environmental group in the present study corroborate with the observation that chromosomes of related organisms generally have similar repeat densities. The exceptions to this indicate differences in repair mechanisms or selection pressures, or both (Achaz et al., 2002), leading to differences in SSR evolution and density in genomes of different sizes (Trivedi, 2004; Lim et al., 2004). This study raises questions about whether an increase in genome complexity should be attributed to SSRs, because, at least in prokaryotes, repeats constitute a low percentage of the total genome (as found here); other factors in addition to tandem repeats may be responsible for the increase in genome size (Hancock, 2002). Genome size and optimum growth temperature correlation with repeat length The genome size was not correlated with average repeat lengths of SSRs in the Archaea. However, the trends to be watched in the future are a positive correlation of total SSRs and trinucleotide average repeat lengths (‘r’ = 0.495 and 0.498; P < 0.05, respectively) and maximum repeat lengths (data not shown; r = 0.4738; P < 0.05) in 19 Archaea. Further studies may confirm whether motif-specific increase in repeat length in Archaea corroborates studies reported for other organisms (Harr et al., 2002). Short repeat lengths were generally found in fungal genome, where long repeats in large genomes are exceptions rather than the rule, also found by Lim et al. (2004). For example, M. acetivorans has the largest genome size, but has a maximum repeat length of 37 bases, but M. mazei, which has the second largest genome size, has a maximum repeat length of 164 bases. Although coding sequences generally have few long repeats (Dokholyan et al., 2000; Morgante et al., 2002), they are present in M. mazei AAT repeat (137 bases) coding sequences of conserved protein, ATA (52 bases) in transcriptional regulator and ArsR family, and AAG (50 bases) in hypothetical protein. Methanobacterium thermoautotrophicum coding sequences have AGC (39 bases) in ribosomal protein, in Halobacterium (CGCAG, 35 bases) and in M. maripaludis (AAAAT) conserved hypothetical protein. There was no correlation of SSR average repeat length with OGT in the 19 Archaea and in the three environmental groups. However, total SSR and pentanucleotide repeats were inversely correlated (‘r’ = -0.513 and -0.574; P < 0.05) in all the Archaea species. Although this correlation was not highly significant, with analysis of more Archaea genomes it may be possible to confirm whether motif-dependent length variations in Archaea are due to differential mutation rates (Toth et al., 2000; Webster et al., 2002). It is also known that SSR motifs, tract type, genomic locations, and selection pressures may influence lengths that are dynamic and may increase in some species, while in others they may decrease (Bowater et al., 1997; Harr et al., 2002). However, from the present study it cannot be concluded whether repeats are unstable (expanding or reducing in size) in Archaea. If there is instability of repeats in Archaea, it could be due to lack of mismatch repair systems (Fitz-Gibbon et al., 2002). Genome CG content, SSR density and SSR CG richness Archaea genomes revealed varying CG richness (Table 1). All repeat types in all Archaea, hyperthermophiles (except dinucleotides) and thermophiles (except di- and tetranucleotides) showed a positive correlation of CG richness of SSRs with CG richness of the genome (Table 5). There was no correlation of genome CG content with total SSR and total motif density, except an inverse correlation of pentanucleotide repeats (‘r’ = -0.621 and -0.604; P < 0.05) in genome and coding regions (‘r’ = -0.832; P< 0.005). However, the trends to be watched in the future are a positive correlation of trinucleotide density in genome and coding regions (‘r’ = 0.493 and 0.524; P < 0.05). Hyperthermophiles had an inverse correlation of total SSR, tetra- and pentanucleotides (‘r’ = -0.6406, -0.6701 and -0.7394, respectively; P < 0.05) in the genome. Trinucleotides in thermophiles showed a positive correlation (‘r’ = 0.8371, 0.8494; P < 0.05) in the total genome and in coding regions. With availability of more Archaea genome sequences, it may be confirmed whether with increasing CG content of the genome, the density of some SSR motifs increases, as in fungal genomes (Lim et al., 2004). It may confirm whether nucleotide composition of the genome influences some repeat types and possibly their generation and amplification (Achaz et al., 2002) in Archaea. Correlation analysis shows that OGT has no influence on CG richness of SSR in total genome, coding and intergenic sequences in all Archaea, and in hyperthermophiles, thermophiles and mesophiles, analyzed separately.
Trends in mesophiles showed an inverse correlation of dinucleotides (‘r’ = -0.9993; P < 0.05) and a positive correlation of pentanucleotides (‘r’ = 0.9999; P < 0.001) with OGT in the total genome. The CG content of genome dinucleotide density showed a positive correlation (‘r’ = 0.9971; P < 0.05) in the total genome. An indication of inverse correlation of OGT with pentanucleotide density (‘r’ = 0.99949; P < 0.05) and tetranucleotide CG richness (‘r’ = 0.99905; P < 0.05) was also seen. Only pentanucleotide CG richness had a positive correlation with genome CG richness. From the foregoing information, it is evident that SSRs are common in Archaea genomes, which may be due to certain advantages to the organisms and may be a “molecular device” for quick adaptations to environmental stress (Young et al., 2000; Li et al., 2002). The variations in distribution, abundance and motif preferences among Archaea could be due to different adaptive strategies, as in other organisms (Rocha et al., 1999). It would therefore be interesting to investigate other stress factors, such as salinity, radiation and pressure, in addition to temperature and presence or absence of efficient repair and replication machinery to understand the reasons for these differences. For example, in Halobacterium, environmental stress may have resulted in a high density of SSRs, which might function as a source for the generation of genetic diversity, allowing the organism to better respond to a wide range of stress, due to salinity, UV radiation, oxygen, and nutrients (Kennedy et al., 2001). CONCLUSIONS It is evident that unlike the other two domains, some Archaea have abundant SSRs in coding regions. There is preferential distribution of repeat motifs, and some motifs are organism specific. Few SSRs are present partially in intergenic and coding sequences. There was no correlation of SSR density with genome size, CG richness of genome or OGT. Similarly, OGT had no influence on CG richness or repeat length of SSRs. Analysis of more sequences from Archaea living in different environmental niches may help us to understand the influence of extreme conditions on SSRs. However, the significance of many of these repeats in Archaea is not known; further investigation may reveal an answer. REFERENCES Achaz G, Rocha EPC, Netter P and Coissac E (2002). Origin and fate of repeats in bacteria. Nucleic Acids Res. 30: 2987-2994. Alba MM, Santibanez-Koref MF and Hancock JM (1999a). Amino acid reiterations in yeast are overrepresented in particular classes of proteins and show evidence of a slippage-like mutational process. J. Mol. Evol. 49: 789-797. Alba MM, Santibanez-Koref MF and Hancock JM (1999b). Conservation of polyglutamine tract size between mice and humans depends on codon interruption. Mol. Biol. Evol. 16: 1641-1644. Alba MM, Santibanez-Koref MF and Hancock JM (2001). The comparative genomics of polyglutamine repeats: extreme difference in the codon organization of repeat-encoding regions between mammals and Drosophila. J. Mol. Evol. 52: 249-259. Bachtrog D, Weiss S, Zangerl B, Brem G, et al. (1999). Distribution of dinucleotide microsatellites in the Drosophila melanogaster genome. Mol. Biol. Evol. 16: 602-610. Bachtrog D, Agis M, Imhof M and Schlotterer C (2000). Microsatellite variability differs between dinucleotide repeat motifs - evidence from Drosophila melanogaster. Mol. Biol. Evol. 17: 1277-1285. Bowater RP, Jaworski A, Larson JE, Parniewski P, et al. (1997). Transcription increases the deletion frequency of long CTG.CAG triplet repeats from plasmids in Escherichia coli. Nucleic Acids Res. 25: 2861-2868. Butcher RD, Hubbard SF and Whitfield WG (2000). Microsatellite frequency and size variation in the parthenogenetic parasitic wasp Venturia canescens (Gravenhorst) (Hymenoptera: Ichneumonidae). Insect Mol. Biol. 9: 375-384. Campbell A, Mrazek J and Karlin S (1999). Genome signature comparisons among prokaryote, plasmid and mitochondrial DNA. Proc. Natl. Acad. Sci. USA 96: 9184-9189. Cardle L, Ramasy L, Milbourne D, Macaulay M, et al. (2000). Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 156: 847-854. Chakravarty S and Varadarajan R (2000). Elucidation of determinants of protein stability through genome sequence analysis. FEBS Lett. 470: 65-69. Chakravarty S and Varadarajan R (2002). Elucidation of factors responsible for enhanced thermal stability of proteins: a structural genomics based study. Biochemistry 41: 8152-8161. Chambers KG and MacAvoy ES (2000). Microsatellites: consensus and controversy. Comp. Biochem. Physiol. (Part B). 126: 455-476. Cleary JD, Nichol K, Wang YH and Pearson CE (2002). Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat. Genet. 31: 37-46. Cox R and Mirkin SM (1997). Characteristic enrichment of DNA repeats in different genomes. Proc. Natl. Acad. Sci. USA 94: 5237-5242. Crollius RH, Jaillon O, Dasilva C, Ozouf-Costaz C, et al. (2000). Characterization and repeat analysis of the compact genome of the fresh water Pufferfish Tetraodon nigroviridis. Genome Res. 10: 939-949. de Farias ST and Bonato MCM (2002). Preferred codons and amino acid couples in hyperthermophiles. Genome Biol. 3 (http://genomebiology.com/2002/3/8/preprint/0006). Dokholyan NV, Buldyrev SV, Havlin S and Stanley HE (2000). Distributions of dimeric tandem repeats in non-coding and coding DNA sequences. J. Theor. Biol. 202: 273-282. Doolittle RF (1995). Of Archaea and Eo: what’s in a name? Proc. Natl. Acad. Sci. USA 92: 2421-2423. Elgar G, Clark MS, Meek S, Smith S, et al. (1999). Generation and analysis of 25 Mb of genomic DNA from the Pufferfish Fugu rubripes by sequence scanning. Genome Res. 9: 960-971. Ellegren H (2000). Heterogeneous mutation processes in human microsatellite DNA sequences. Nat. Genet. 24: 400-402. Farias ST and Bonato MC (2003). Preferred amino acids and thermostability. Genet. Mol. Res. 2: 383-393. http://www.funpecrp.com.br/gmr. Fitz-Gibbon ST, Ladner H, Kim U, Stetter KO, et al. (2002). Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. USA 99: 984-989. Gentles AJ and Karlin S (2001). Genome-scale compositional comparisons in eukaryotes. Genome Res. 11: 540-546. Godde JS and Wolffe AP (1996). Nucleosome assembly on CTG triplet repeats. J. Biol. Chem. 271: 15222-15229. Grayling RA, Bailey KA and Reeve JN (1997). DNA binding and nuclease protection by the HMf histones from the hyperthermophilic archaeon Methanothermus fervidus. Extremophiles 1: 79-88. Green H and Wang N (1994). Codon reiteration and evolution of proteins. Proc. Natl. Acad. Sci. USA 91: 4298-4302. Gromiha MM, Thomas S and Santhosh C (2002). Role of cation-pi interactions to the stability of thermophilic proteins. Prep. Biochem. Biotechnol. 32: 355-362. Gur-Arie R, Cohen CJ, Eitan Y, Shelef L, et al. (2000). Simple sequence repeats in Escherichia coli: abundance distribution composition and polymorphism. Genome Res. 10: 62-71. Hancock JM (1996a). Simple sequences and the expanding genome. Bioessays 18: 421-425. Hancock JM (1996b). Simple sequences in a “minimal” genome. Nat. Genet. 14: 14-15. Hancock JM (2002). Genome size and accumulation of simple sequence repeats: implications of new data from genome sequencing projects. Genetica 115: 93-103. Hancock JM and Santibanez-Koref MF (1998). Trinucleotide expansion diseases in the context of micro- and minisatellite evolution. EMBO J. 17: 5521-5524. Harr B, Zangerl B, Brem G and Schlotterer C (1998). Conservation of locus specific microsatellite variability across species’ a comparison of two Drosophila sibling species D. melanogaster and D. simulans. Mol. Biol. Evol. 15: 176-184. Harr B, Zangerl B and Schlotterer C (2000). Removal of microsatellite interruptions by DNA replication slippage: phylogenetic evidence from Drosophila. Mol. Biol. Evol. 17: 1001-1009. Harr B, Todorova J and Schlotterer C (2002). Mismatch repair-driven bias in D. melanogaster. Mol. Cell. 10: 199-205. Hartenstine MJ, Goodman MF and Petruska J (2000). Base stacking and even/odd behavior of hairpin loops in DNA triplet repeat slippage and expansion with DNA polymerase. J. Biol. Chem. 275: 18382-18390. Hashem VI, Rosche WA and Sinden RR (2002). Genetic assays for measuring rates of (CAG)·(CTG) repeat instability in Escherichia coli. Mutat. Res. 502: 25-37. Hickey A and Singer GAC (2004). Genomic and proteomic adaptations to growth at high temperature. Genome Biol. 5: 117.1-117.7. Ireland MJ, Reinkea SS and Livingston DM (2000). The impact of lagging strand replication mutations on the stability of CAG repeat tracts in yeast. Genetics 155: 1657-1665. Jakupciak JP and Wells RD (2000). Gene conversion (recombination) mediates expansions of CTG·CAG repeats. J. Biol. Chem. 275: 40003-40013. Karlin S, Mrazek J and Campbell AM (1997). Compositional biases of bacterial genomes and evolutionary implications. J. Bacteriol. 179: 3899-3913. Katti MV, Ranjekar PK and Gupta VS (2001). Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol. Biol. Evol. 18: 1161-1167. Kennedy SP, Ng WV, Salzberg SL, Hood L, et al. (2001). Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 11: 1641-1650. Lerat E, Capy P and Biemont C (2002). The relative abundance of dinucleotides in transposable elements in five species. Mol. Biol. Evol. 19: 964-967. Li FY, Leibiger B, Leibigerand I and Larsson C (2002). Characterization of a putative murine mitochondrial transporter homology of hMRS3/4. Mamm. Genome 13: 20-23. Li Y, Korol AB, Fahima T and Nevo E (2004). Microsatellites within genes: structure, function and evolution. Mol. Biol. Evol. 21: 991-1007. Lim S, Notley-McRobba L, Lim M and Carter DA (2004). A comparison of the nature and abundance of microsatellites in 14 fungal genomes. Fungal Genet. Biol. 41: 1025-1036. Makarova KS and Koonin EV (2003). Comparative genomics of Archaea: how much have we learned in six years and what’s next? Genome Biol. 4: 115.1-115.17. Marcotte EM, Pellegrini M, Yeates TO and Eisenberg D (1999). A census of protein repeats. J. Mol. Biol. 293: 151-160. Metzgar D, Bytof J and Wills C (2000). Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 10: 72-80. Mishima Y, Kaizu H and Kominami R (1997). Pairing of DNA fragments containing (GGA.TCC)n repeats and promotion by high mobility group protein 1 and histone H1. J. Biol. Chem. 272: 26578-26584. Moore H, Greenwell PW, Liu CP, Arnheim N, et al. (1999). Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl. Acad. Sci. USA 96: 1504-1509. Morel P, Reverdy C, Michel B, Ehrlich SD, et al. (1998). The role of SOS and flap processing in microsatellite instability in Escherichia coli. Proc. Natl. Acad. Sci. USA 95: 10003-10008. Morgante M, Hanafey M and Powell W (2002). Microsatellites are preferentially associated with nonrepetitive DNA in plant genome. Nat. Genet. 30: 194-200. Morris J, Kushner SR and Ivarie R (1986). The simple repeat poly(dT-dG).poly(dC-dA) common to eukaryotes is absent from eubacteria and archaebacteria and rare in protozoans. Mol. Biol. Evol. 3: 343-355. Ogura T, Okano K, Tsuchida K, Miyajima N, et al. (1994). A defective non-LTR retrotransposon is dispersed throughout the genome of the silkworm Bombyx mori. Chromosoma 103: 311-323. Okonogi TM, Alley SC, Reese AW, Hopkins PB, et al. (2000). Sequence-dependent dynamics in duplex DNA. Biophys. J. 78: 2560-2571. Pereira SL and Reeve JN (1998). Histones and nucleosomes in Archaea and Eukarya: a comparative analysis. Extremophiles 2: 141-148. Pereira SL, Grayling RA, Lurz R and Reeve JN (1997). Archaeal nucleosomes. Proc. Natl. Acad. Sci. USA 94: 12633-12637. Primmer CR and Ellegren H (1998). Patterns of molecular evolution in avian microsatellites. Mol. Biol. Evol. 15: 997-1008. Primmer CR, Raudsepp T, Chowdhary BP, Moller AP, et al. (1997). Low frequency of microsatellites in the avian genome. Genome Res. 7: 471-482. Ramsay L, Macaulay M, Delgi Ivanissevich S, MacLean K, et al. (2000). A simple sequence repeat-based linkage map of barley. Genetics 156: 1997-2005. Reeve JN, Sandman K and Daniels CJ (1997). Archaeal histones, nucleosomes and transcription initiation. Cell 89: 999-1002. Rocha EPC and Blanchard A (2002). Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 30: 2031-2042. Rocha EPC, Danchin A and Viari A (1999). Analysis of long repeats in bacterial genomes reveals alternative evolutionary mechanisms in Bacillus subtilis and other competent prokaryotes. Mol. Biol. Evol. 16: 1219-1230. Rocha EPC, Maticand I and Taddei F (2002). Over-representation of repeats in stress response genes: a strategy to increase versatility under stressful conditions? Nucleic Acids Res. 30: 1886-1894. Sandman K and Reeve JN (2000). Structure and functional relationships of archaeal and eukaryal histones and nucleosomes. Arch. Microbiol. 173: 165-169. Sandman K and Reeve JN (2001). Chromosome packaging by archaeal histones. Adv. Appl. Microbiol. 50: 75-99. Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, et al. (2000). Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 97: 5750-5755. Schlotterer C (1998). Genome evolution: are microsatellites really simple sequences? Curr. Biol. 8: R132-R134. Schlotterer C (2000). Evolutionary dynamics of microsatellite DNA. Chromosoma 109: 365-371. Smith SC, Kennelly PJ and Potts M (1997). Protein-tyrosine phosphorylation in the Archaea. J. Bacteriol. 197: 2418-2420. Stallings RL (1995). Conservation and evolution of (CT)n/(GA)n microsatellite sequences at orthologous positions in diverse mammalian genomes. Genomics 25: 107-113. Subramanian S, Madgula VM, George R, Mishra RK, et al. (2003). Triplet repeats in human genome: distribution and their association with genes and other genomic regions. Bioinformatics 19: 549-552. Thiel T, Michalek W, Varshney K and Graner A (2003). Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106: 411-422. Toth G, Gaspari Z and Jurka J (2000). Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 10: 967-981. Trivedi S (2003). Do microsatellites have biased associations? Nucleus 46: 61-76. Trivedi S (2004). Microsatellites (SSRs): puzzles within puzzles. Ind. J. Biotech. 3: 331-347. Varshney RK, Thiel T, Stein N, Langridge P, et al. (2002). In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Ann. Neurol. 52: 498-503. Wang Y, Rocha EP, Leung FC and Danchin A (2004). Cytosine methylation is not the major factor inducing CpG dinucleotide deficiency in bacterial genomes. J. Mol. Evol. 58: 692-700. Webster MT, Smith NG and Ellegren H (2002). Microsatellite evolution inferred from human-chimpanzee genomic sequence alignments. Proc. Natl. Acad. Sci. USA 99: 8748-8753. Woese CR, Kandler O and Wheelis ML (1990). Towards a natural system of organisms: proposal for the domain Archaea, Bacteria and Eucarya. Proc. Natl. Acad. Sci. USA 87: 4576-4579. Wren JD, Forgacs E, Fondon JW 3rd, Pertsemlidis A, et al. (2000). Repeat polymorphisms within gene regions: phenotypic and evolutionary implications. Am. J. Hum. Genet. 67: 345-356. Young ET, Sloana JS and Riper KV (2000). Trinucleotide repeats are clustered in regulatory genes in Saccharomyces cerevisiae. Genetics 154: 1053-1068. Zhu Y, Strassmann JE and Queller D (2000). Insertions substitutions and origin of microsatellites. Genome Res. 76: 227-236. Zlatanova J (1997). Archaeal chromatin: virtual or real? Proc. Natl. Acad. Sci. USA 94: 12251-12254.
|
|