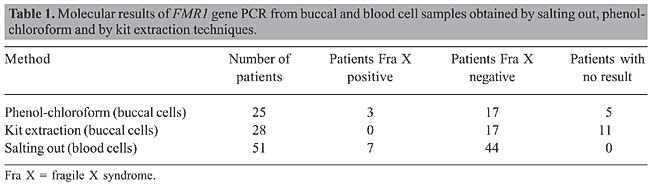
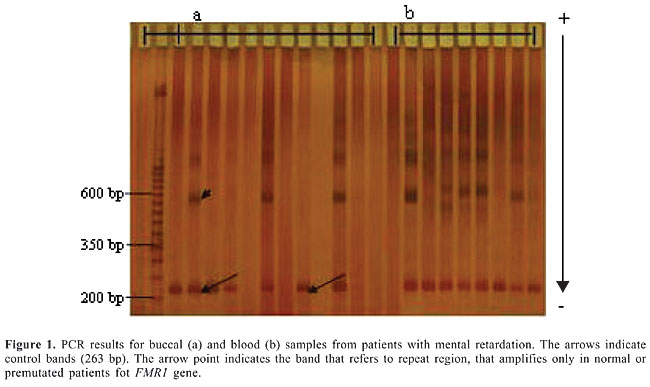
ABSTRACT. Fragile X syndrome is one of the most frequent causes of mental retardation. Since the phenotype in this syndrome is quite variable, clinical diagnosis is not easy and molecular laboratory diagnosis is necessary. Usually DNA from blood cells is used in molecular tests to detect the fragile X mutation which is characterized by an unstable expansion of a CGG repeat in the fragile X mental retardation gene (FMR1). In the present study, blood and buccal cells of 53 mentally retarded patients were molecularly analyzed for FMR1 mutation by PCR. Our data revealed that DNA extraction from buccal cells is a useful noninvasive alternative in the screening of the FMR1 mutation among mentally retarded males. Key words: Fragile X syndrome, Mental retardation, Buccal cells, DNA extraction INTRODUCTION Mental retardation is one of the most common human disorders. It may appear isolated or as one of the clinical signs of a more complex syndrome as in Down syndrome and in fragile X syndrome (Toniolo, 2000). Fragile X syndrome (FRAXA - MIM 309550) is the most frequent inherited form of mental retardation in males and affects 1 in 4,000-6,000 males and 1 in 7,000-10,000 females (Crawford et al., 2001). This syndrome results from a dynamic mutation characterized by an abnormal expansion of a trinucleotide (CGG) repeat located in the 5’-untranslated region in the first exon of the fragile X mental retardation gene (FMR1) (Fu et al., 1991; Verkerk et al., 1991). Expansion of the CGG repeat beyond 200 trinucleotides results in hypermethylation of both the FMR1 promoter region and CGG repeat. As a consequence, the gene is transcriptionally silenced, and there is no gene product (FMRP) in affected individuals (Pieretti et al., 1991; Sutcliffe et al., 1992). The absence of FMRP in the brain is responsible for cognitive impairment (Verheij et al., 1993). In males, the full mutation is associated with mild to severe mental retardation, whereas in females around 60% with full mutation present mild to moderate mental impairment, considering random X inactivation (Willemsen et al., 2003). Even in males, fragile X patient phenotypes are quite variable. The main clinical feature is mental retardation, ranging from mild learning disability to profound mental retardation with frequent occurrences of autistic-like behavior and hyperactivity. Physical and behavioral characteristics are variably expressed depending on age and sex (Hagerman et al., 1991). Suggestive clinical signs, such as mild facial dysmorphia with long face and large ears, and macroorchidism established around puberty, are not sufficiently consistent or specific to establish or exclude diagnosis (Mandel and Biancalana, 2004). Thus, fragile X syndrome is difficult to ascertain clinically, requiring molecular laboratory diagnosis. Although blood samples are the best choice for large amounts of genomic DNA, the collection of peripheral blood is not easy in some patients, especially in children and syndromic patients, who may show behavioral difficulties. Different methods of DNA extraction from other tissues have been described in the literature, such as from hair root (Higuchi et al., 1988) and buccal epithelium (Moore et al., 2001; Zheng et al., 2001). Buccal cell collection provides a noninvasive method for obtaining DNA (Zheng et al., 2001). Exfoliated buccal epithelium cells are a promising alternative source of DNA since it can be easily obtained using relatively inexpensive techniques. Buccal swabs and mouthwash protocols are most often used for buccal cell collection (Garcia-Glosas et al., 2001). The present study describes the use of blood and buccal cell samples for extraction of DNA used in PCR for the detection of the fragile X syndrome among patients with mental retardation. MATERIAL AND METHODS Subjects Fifty-three male patients with idiopathic mental retardation were included in this study. Patients’ age ranged from 3 to 31 years. Patients with karyotypic aberration using G-banding or some other known cause of mental retardation were not included in this study. Samples were obtained after patients’ informed consent and approval by UNIFESP’s Ethics Committee. Blood and buccal cell collection and DNA extraction Peripheral blood (5 mL) was collected from each patient. DNA samples were obtained by a salting-out method modified from Lahiri and Nurnberger Jr. (1991). Buccal cells were collected with a cytobrush. The patients were instructed to brush their teeth and refrain from eating and drinking for at least 1 h before sample collection. Patients had each cheek brushed for at least 30 s and the brushes were placed in plastic tubes. DNA from buccal cells was obtained using two different methods: one using phenol-chloroform extraction modified from Garcia-Glosas et al. (2001), and one using a DNA purification kit (Amershan GFX Genomic blood, Amershan Pharmacia Biotech Inc.®, USA). For phenol-chloroform extraction, 700 µL lysis buffer (0.01 M Tris-HCl, 0.01 M EDTA, 0.1 M NaCl, 2% SDS) was added to the plastic sample tubes, and the tubes were mixed and incubated for 10 min at room temperature. Next, 35 µL of proteinase K was added to the suspension, and the tubes were mixed and incubated at 58°C for 2 h. Afterward, the samples were centrifuged at 12,000 rpm for 15 min. The cytobrush was removed and an equal volume of saturated phenol was added. The tubes were then mixed for 10 s and centrifuged at 12,000 rpm for 5 min. The aqueous layer was transferred to a new tube and 1 mL of phenol-chloroform-isoamyl alcohol (25:24:1) was added. The tubes were then mixed for 10 s and centrifuged at 12,000 rpm for 5 min. This step was repeated one more time. The aqueous layer was transferred to a new tube and two volumes of 100% ethanol and 0.1 volume of 3 M NaOAc, pH 6.0, were added. Samples were stored overnight at -20°C and then centrifuged at 12,000 rpm for 20 min. The DNA pellet was washed with 70% ethanol, dried for 15 min, resuspended in 25 µL of water and stored at 4°C. DNA extraction using a DNA purification kit (Amershan GFX Genomic blood®) was performed following the protocol for purification of buccal cells. The cytobrush was placed in a 1.5-mL microcentrifuge tube. Next, 500 µL of extraction solution was added to the sample, and the tubes were mixed and incubated for 10 min at room temperature. The extraction mixture was transferred to the GFX column placed in a collection tube which was centrifuged at 5,000 g for 1 min. Next, 500 µL of extraction solution was added to the column and centrifuged at 5,000 g for 1 min. The effluent was discarded by emptying the collection tube. Afterward, 500 µL of wash solution was added to the column and centrifuged at full speed for 3 min. The column was placed in a fresh tube and 200 µL of pre-heated elution buffer was directly applied to the glass fiber in the GFX column, incubated for 1 min and centrifuged at 5,000 g for 1 min to recover purified DNA. Molecular analysis DNA samples were analyzed by PCR according to Haddad et al. (1996). In this method, three primers that amplify a control band and a band which includes the CGG repeat (with up to 78% CG content) were used. Fragile X patients showed only the control band amplified since the DNA segment with a large number of CGG failed to amplify under our experimental conditions. PCR samples were analyzed using GeneGel Excel acrylamide gels (GE Health Care®) and silver nitrate staining. Three reactions were performed on each patient sample to confirm the results. We evaluated different methods of DNA extraction considering quality and quantity of the material obtained and procedure feasibility. RESULTS Molecular evaluation PCR molecular analysis from blood samples was possible for 51 patients. Two patients became aggressive and anxious making collection impossible. Seven patients were positive for fragile X syndrome and the others were normal for FMR1 gene. Buccal cells were obtained from all 53 patients. In 25 patients, DNA was extracted from buccal cells using phenol-chloroform method. From these patients, we obtained PCR results in 20 cases, 17 normal for FMR1 and three with FMR1 mutation. In 28 patients, we used a commercial kit for DNA extraction, obtaining PCR results in 17 of them, all normal for FMR1 mutation (Table 1). Buccal cell DNA successfully yielded PCR results, and there was full agreement with blood sample findings (Figure 1).
DISCUSSION Considering the epidemiological importance of fragile X syndrome, a good screening method involving a simple, fast and inexpensive technique is desirable (Mingroni-Netto et al., 1996; Haddad et al., 1996). The discovery of the fragile X expansion mutation has produced efficient and reliable tools for diagnosis, genetic counseling and prenatal diagnosis (Rousseau et al., 1991). The mutation is generally tested by the Southern blot method, which allows an evaluation of both the expansion and gene methylation status. PCR-based methods are also useful, especially for precisely sizing premutations or for excluding a diagnosis of fragile X, when a normal CGG repeat allele is found in a male patient (Mandel and Biancalana, 2004). However, DNA diagnosis is difficult in patients with mental retardation due to recalcitrance to blood drawing. Therefore, an alternative and less invasive method of DNA collection is important for these patients, and extraction from buccal cells is convenient. Comparing the three methods used in this study, DNA extraction from blood showed the best quality and quantity. DNA extraction from buccal cells resulted in a smaller number of cases with sufficient DNA for molecular testing. DNA extraction from phenol-chloroform provided good-quality material. PCR amplification proved to be impossible in 20% of the cases (5/25). On the other hand, DNA extraction using a commercial kit, although quicker and easier, resulted in a lower amount of DNA, where amplification failed in 39% of samples (11/28). Amplification of buccal cell DNA allowed the discrimination between affected and unaffected full mutation males. Band quality observed in acrylamide gels was similar to that for lymphocyte DNA demonstrating equivalent results (Figure 1). FMR1 gene analysis by PCR can be performed on small amounts of blood and other tissues. Hagerman et al. (1994) described a pilot program using saliva testing for the FMR1 mutation in children with learning disabilities and found agreement between buccal and blood cells. Our results showed that although buccal cell DNA extraction results in a lower amount of DNA than with lymphocytes it is a good technical alternative for diagnostic purposes. The limited success of PCR from buccal cells could be attributed to insufficient DNA obtained from children with the use of cytobrushes (Zheng et al., 2001). The advantages of buccal cell DNA for fragile X syndrome screening in males are: 1) obtaining the material is easy and painless, two important considerations when dealing with retarded patients; 2) the patient’s mother or other relative can collect the material, even at home, just requiring a tube with saline and a cytologic brush, and 3) test results are reliable as a screening test. Thus, PCR testing using buccal cell DNA is considered a helpful screening tool for male patients with idiopathic mental retardation. ACKNOWLEDGMENTS Research supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil. REFERENCES Crawford DC, Acuna JM and Sherman SL (2001). FMR1 and the fragile X syndrome: human genome epidemiology review. Genet. Med. 3: 359-371. Fu YH, Kuhl DP, Pizzuti A, Pieretti M, et al. (1991). Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 67: 1047-1058. Garcia-Glosas M, Egan K, Abruzzo J, Newcomb P, et al. (2001). Collection of genomic DNA from adults in epidemiological studies by buccal cytobrushes and mouthwash. Cancer Epidemiol. Biomarkers Prev. 10: 687-696. Haddad LA, Mingroni-Netto RC, Vianna-Morgante AM and Pena SD (1996). A PCR-based test suitable for screening for fragile X syndrome among mentally retarded males. Hum. Genet. 97: 808-812. Hagerman RJ, Amiri K and Cronister A (1991). Fragile X checklist. Am. J. Med. Genet. 38: 283-287. Hagerman RJ, Wilson P, Staley LW, Lang KA, et al. (1994). Evaluation of school children at high risk for fragile X syndrome utilizing buccal cell FMR-1 testing. Am. J. Med. Genet. 51: 474-481. Higuchi R, von Beroldingen CH, Sensabaugh GF and Erlich HA (1988). DNA typing from single hairs. Nature 332: 543-546. Lahiri DK and Nurnberger Jr JI (1991). A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 19: 5444. Mandel JL and Biancalana V (2004). Fragile X mental retardation syndrome: from pathogenesis to diagnostic issues. Growth Horm. IGF Res. 14 (Suppl A): S158-S165. Mingroni-Netto RC, Haddad LA and Vianna-Morgante AM (1996). The number of CGG repeats of the FMR1 locus in premutated and fully mutated heterozygotes and their offspring: implications for the origin of mosaicism. Am. J. Med. Genet. 64: 270-273. Moore L, Wiencke J, Eng C, Zheng S, et al. (2001). Evaluation of buccal cell collection protocols for genetic susceptibility studies. Biomarkers 6: 448-454. Pieretti M, Zhang FP, Fu YH, Warren ST, et al. (1991). Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66: 817-822. Rousseau F, Heitz D, Biancalana V, Blumenfeld S, et al. (1991). Direct diagnosis by DNA analysis of the fragile X syndrome of mental retardation. N. Engl. J. Med. 325: 1673-1681. Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, et al. (1992). DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet. 1: 397-400. Toniolo D (2000). In search of the MRX genes. Am. J. Med. Genet. 97: 221-227. Verheij C, Bakker CE, de Graaff E, Keulemans J, et al. (1993). Characterization and localization of the FMR-1 gene product associated with fragile X syndrome. Nature 363: 722-724. Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, et al. (1991). Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65: 905-914. Willemsen R, Smits A, Severijnen LA, Jansen M, et al. (2003). Predictive testing for cognitive functioning in female carriers of the fragile X syndrome using hair root analysis. J. Med. Genet. 40: 377-379. Zheng S, Ma X, Buffler PA, Smith MT, et al. (2001). Whole genome amplification increases the efficiency and validity of buccal cell genotyping in pediatric populations. Cancer Epidemiol. Biomarkers Prev. 10: 697-700. |
|