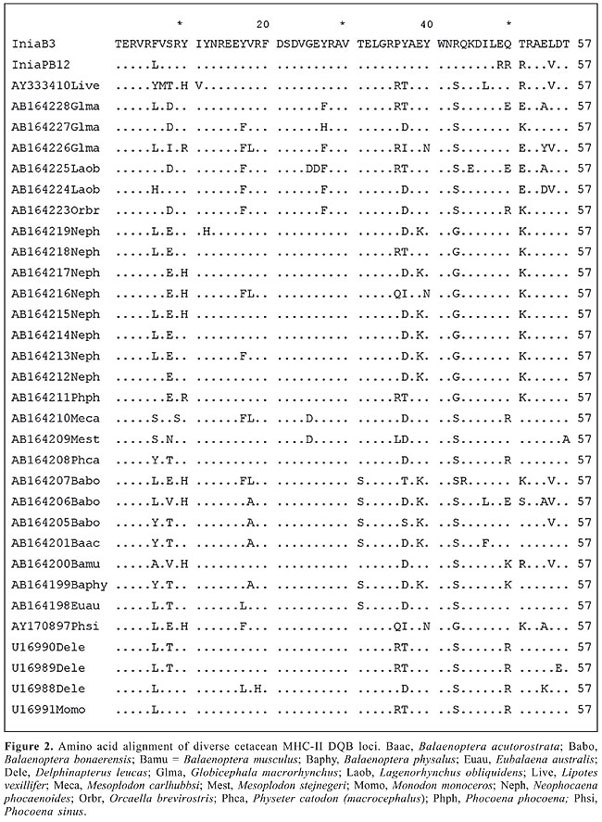
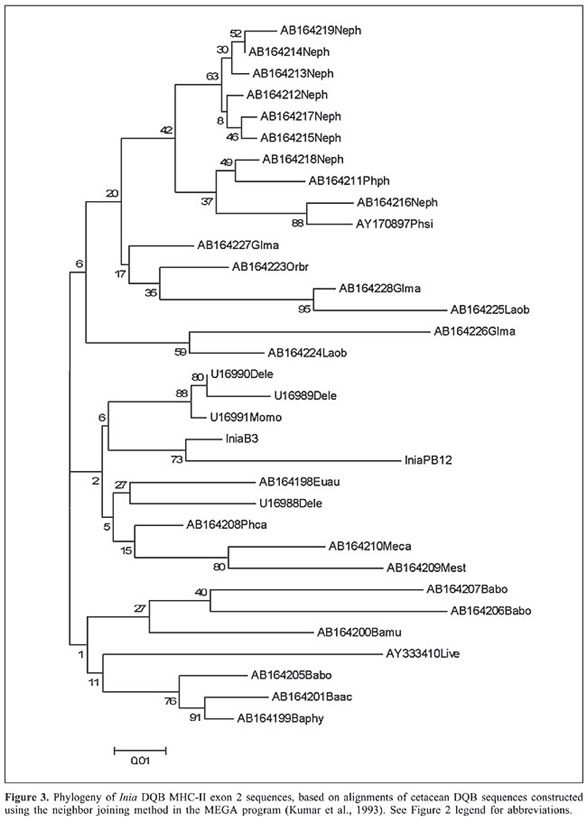
ABSTRACT. We report the first major histocompatibility complex (MHC) DQB1 sequences for the two species of pink river dolphins (Inia geoffrensis and Inia boliviensis) inhabiting the Amazon and Orinoco River basins. These sequences were found to be polymorphic within the Inia genus and showed shared homology with cetacean DQB-1 sequences, especially, those of the Monodontidae and Phocoenidae. On the other hand, these sequences were shown to be divergent from those described for other riverine dolphin species, such as Lipotes vexillifer, the Chinese river dolphin. Two main conclusions can be drawn from our results: 1) the Mhc DQB1 sequences seem to evolve more rapidly than other nuclear sequences in cetaceans, and 2) differential positive selective pressures acting on these genes cause concomitant divergent evolutionary histories that derive phylogenetic reconstructions that could be inconsistent with widely accepted intertaxa evolutionary relationships elucidated with other molecular markers subjected to a neutral dynamics. Key words: Mhc DQB1 sequences, Inia, Molecular evolution, River dolphins INTRODUCTION All gnatostomates (jawed vertebrates) possess major histocompatibility complex (Mhc) genes that encode proteins responsible for binding and displaying self and foreign peptides to T-cells to either eliminate self-reactive T-cells early in development in the thymus or elicit immune responses against pathogens (Doherty and Zinkernagel, 1975; Klein and Takahata, 1990). Specifically, Mhc class II DQB genes are expressed only by antigen-presenting cells, and encode cell surface proteins with a peptide-binding region (PBR) that bind and present self and foreign peptides to CD4 T-cells, to trigger the immune response (Brown et al., 1993). DQB genes comprise 6 exons. Exon 1 encodes the leader peptide, and exon 2, part of the variable PBR. This region is of biological importance because this is where the rest of the polypeptide chain (between 13 to 18 amino acids) of the foreign peptide is linked (Brown et al., 1988). Most of the polymorphism in the Mhc molecules occurs at the PBR, creating variation for binding specificity (Edwards and Hedrick, 1998). Additionally, exons 3, 4 and 5 encode the extracellular, transmembrane and cytoplasmic domains, respectively (Brown et al., 1993). A substantial amount is known about Mhc polymorphism and evolution in humans and certain captive primate populations (e.g., Klein et al., 1993; Bergström et al., 1999; Bontrop et al., 1999). In humans, particular Mhc variants are related to disease resistance (Hill et al., 1991; Jepson et al., 1997; Hill, 1999; Carrington et al., 1999). Such studies suggest that species with low Mhc variation may be more susceptible to infectious disease, and that some endangered species living in small, isolated populations could face additional threats of extinction from exposure to pathogens and parasites (Evermann et al., 1988; Lyles and Dobson, 1993; Mikkos et al., 1999; Murray et al., 1999; Lukas et al., 2004). Mhc polymorphism results from high-nonsynonymous nucleotide substitution rates, which shuffle the amino acids of histocompatible proteins, and ultimately shift the physiochemical properties of Mhc proteins. Such high-nonsynonymous substitution rates are mostly attributed to positive selection, like that exerted by pathogen diversity, load and virulence (Hughes and Nei, 1989; Garrigan and Hedrick, 2003). Additionally, a group of functional genes known to be more polymorphic within the Mhc has been demonstrated to be related to male sexual discrimination on the part of females. For instance, Potts et al. (1991) and Manning et al. (1992) performed different experiments demonstrating that Mhc genotypes can affect mate choice prior to fertilization in rodents. These mammals can distinguish Mhc genotypes on the basis of differential odors in the urine. The first study showed that female mice significantly preferred to settle in areas held by a male with unlike Mhc genotypes. Moreover, females tended to search for males that had Mhc genotypes different from that of the males of her own territory for extraterritorial copulations. The second study demonstrated that mice were able to distinguish full-siblings from half-siblings on the basis of Mhc genotypes (Mhc genotypes as a signal for direct kin recognition). Additionally, the same study could demonstrate that mice were capable of dispensing altruistic behavior depending on the Mhc genotypes of the individuals. In fact, Wedekind et al. (1995) showed that the human females gave higher marks for men’s odors for pleasantness, to men with Mhc genotypes dissimilar to their own (HLA-A, HLA-B and HLA-DR). It seems that if women were to choose a mate whose Mhc genotype is different than her own, they would enjoy better fittness as a result of higher rates of offspring survival and reduced inbreeding (Potts and Wakeland, 1993). On the other hand, Ober et al. (1992) showed in a Hutterite community of South Dakota that when human couples shared alleles at HLA-DR and HLA-B, they had lower fertility and higher spontaneous abortion rates than couples with no shared Mhc alleles. Mhc DQB genes have been used as molecular markers in studies of population genetics, evolution, molecular ecology, and conservation genetics because they are the most polymorphic group of functional genes known from vertebrate genomes (Yuhki and O’Brien, 1990; Hambuch and Lacey, 2002) and because they are submitted to the direct influence of natural selection. The marine mammals are considered a group of special interest within evolutionary genetics due to their special characteristics since they reflect a fast adaptation, in evolutionary time, to an aquatic lifestyle (Milinkovitch and Thewissen, 1997; Heyning and Lento, 2002). This supposes important morphological and physiological modifications and implies that these animals could be subjected to different pathogen pressures compared to those that afflict terrestrial mammals. Most research on cetacean Mhc diversity and evolution is in an exploratory phase (e.g., Trowsdale et al., 1989; Flores-Ramírez et al., 2000, 2004). First assessments of Mhc polymorphism in marine mammals reported low variation in southern elephant seals (Mirounga leonina) and rorquals (Balaenoptera physalus and B. borealis) using MHC restriction fragment length polymorphisms, and were attributed to a supposed weak pathogenic pressure (Trowsdale et al., 1989; Slade, 1992). Later sequencing analyses revealed considerable levels of Mhc-I and Mhc-II polymorphism, due to frequent nonsynonomous substitutions, in beluga whales (Delphinapterus leucas), narwhal (Monodon monoceros), four species of pinnipeds, and gray whales (Eschrichtius robustus) (Murray et al., 1995; Murray and White, 1998; Hoelzel et al., 1999; Flores-Ramírez et al., 2000, 2004), suggesting that Darwinian selection drives Mhc polymorphism in marine mammals, and like for terrestrial vertebrates, this might reflect on their population dynamics (Yuhki and O’Brien, 1990). Only two published studies have been performed at the population level in cetaceans, comprising beluga whales (Delphinapterus leucas) and narwhals (Monodon monoceros) (Murray et al., 1995; Murray and White, 1998). Therefore, Mhc analyses on aquatic mammals have concentrated on marine species. Until now, there is only one published paper on a freshwater mammal. This is the case of the Chinese river dolphin (Yang et al., 2005). We report herein the first sequences of Mhc class II (DQB-1 locus) for the Amazon pink river dolphins (Inia geoffrensis and Inia boliviensis). Some features of this dolphin genus are as follows: the pink river dolphin, also called boto, bufeo or bugeo, has a unique natural history. Therefore, it is noteworthy to explore the significance of Mhc diversity in cetaceans living in freshwater Rivers at the Amazon and Orinoco basins. The genus Inia contains two species, Inia boliviensis, which inhabits the Bolivian Amazon (Mamoré and Guaporé (= Iténez) Rivers and effluents), and Inia geoffrensis, living in the main rivers of the Amazon in Ecuador, Peru, Colombia, and Brazil, and in the Orinoco River basin in Colombia and Venezuela (Banguera-Hinestroza et al., 2002). These two species are separated by 400 km of falls in the Madeira River (from Guayaramerín, in Bolivia, to Porto Velho, in Brazil). Among the river dolphins, the boto seems to have the most stable population sizes, although its IUCN status is vulnerable, as its habitat is over exploited (Best and da Silva, 1989). In spite of Inia’s extensive distribution, migratory patterns and reproductive behavior in general are not known, but certainly are very dynamic given the strong seasonal fluctuations of river margins which are surpassed during the rainy season and expand to cover thousands of kilometers of forest. In contrast, during the dry season, river margins and tributaries retract and the dolphins can be found isolated in small lagoons (Best and da Silva, 1989; Da Silva, 1994). Interestingly, Martin and da Silva (2004a,b) found evidence that adult dolphins are largely segregated by sex and reproductive status, except in low water. Thus, as water levels increase, females and calves move into the most remote parts of the inundated areas, while males move to the main channels and rivers. The overall sex ratio seems to be around one. There are two different kinds of rivers in the Amazon and Orinoco basins. The black rivers, with low productivity because of their low pH range, a high concentration of tannins and few organic resources, and the white rivers, which have contrastingly high levels of suspended sediments and organic resources with high productivity. Formerly, it was believed that dolphins could not live in black rivers due to the low pH. However, recent studies documented the presence of dolphins in such rivers in relation to fish density and not to water pH. Thus, these animals live in two different habitats and probably the pathogen pressure on these dolphins in these two different river systems is different as well. In summary, two Inia species appear to occupy quite distinct habitats separated by 400 km of inaccessible habitat dominated by waterfalls. Both habitats are intrinsically heterogeneous, displaying seasonal flooding and specific ecological settings in specific black or white tributaries. The latter has compelled us to hypothesize that dolphins inhabiting such divergent habitats, as those of the main and Bolivian Amazon basins, have been subjected to differential Darwinian selection associated with their distinct parasite diversity, virulence and load, which have shaped the evolution and polymorphism of their pathogen recognition system. As a consequence, we conducted this preliminary study aiming to isolate and characterize the first Mhc DQB-1 sequences from Inia, which would lead to future studies to elucidate the significance of Mhc polymorphism in phylogenetically related cetacean species under distinct selection regimes. Finally, studies are scarce due to the almost impossible task of taking blood samples from cetaceans without disturbing and compromising the welfare of these highly protected or endangered animals, as with the river dolphins. MATERIAL AND METHODS To isolate and characterize Inia’s Mhc DQB-1 sequences, skin biopsies were obtained from two individuals, one I. geoffrensis (PB12) and one I. boliviensis (B3), that were respectively sampled in Peruvian and Bolivian Amazon tributaries. The exact geographic locations for these animals were as follows. The Peruvian dolphin was caught at Tipishca del Loro in the Curaray River. It was an adult male of 195 cm. The Bolivian specimen was an adult male 207 cm in length and was caught at the Porvenir Lagoon in the Mamore River. In the laboratory, total DNA was extracted from each sample using a standard organic extraction protocol (Sambrook et al., 1989). Primers previously designed to amplify human and homologous cetacean DQB-1 sequences (Murray et al., 1995; Munguía-Vega, 2002): 5’-CTG GTA GTT GTG TCT GCA CAC-3' and 5’-CAT GTG CTA CTT CAC CAA CGG-3', were used to amplify 172 bp of the variable DQB-1 exon 2 that encode the PBR and thus most of the functional residues of class II DQB molecules. PCR products of expected size were obtained using 0.5 mM of each primer, 1X buffer, 2.5 mM MgCl2, 1 U Taq polymerase (Invitrogen), 0.2 mM dNTPs, and 10-50 ng DNA. PCR reactions were conducted in a Techne Genius thermocycler, according to a thermal profile consisting of 1 cycle at 94°C for 1 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min, with a final extension at 72°C for 10 min. PCR products of expected size were cloned (pCR2,1 vector, Topo TA cloning kit, Invitrogen) and sequenced on both strands (Macrogene Inc., Seoul, Korea). The histocompatibility nature of the sequences obtained was confirmed based on their shared homology with vertebrate DQB-1 sequences deposited in GeneBank, using the NCBI BLAST search tool. The sequences were aligned using Clustal W (Thompson et al., 1994). A gene tree analysis of boto DQB sequences was performed with the neighbor joining method (Saitou and Nei, 1987) using the Kimura’s two-parameter distance (Kimura, 1980), and with the help of MEGA 2.1 software (Kumar et al., 1993). RESULTS AND DISCUSSION The two DQB DNA sequences obtained are shown in Figure 1 and were deposited in the GeneBank. The deduced amino acid translation of the sequences produced two different proteins with open-reading frames (Figure 2) that seem to be functional. A comparison with the GeneBank database sequences found that Inia DQB exon 2 sequences share high similarity with other Odontoceti, such as sperm whales (Physeteridae) and Monodontidae, while the amino acid translation sequence showed similarity with sperm whale and porpoise (Phocoenidae) sequences. Figure 3 shows the phylogenetic relationships among the cetacean sequences analyzed. Bootstrap values supported clustering between Inia and the Monodontidae (narwhales and beluga whales), one of the families of Delphinoidea. Maximum parsimony trees were also constructed; they showed the same topology as the tree presented in Figure 3.
These results could be compared with some previously published cetacean phylogenies incorporating several river dolphins (Milinkovitch et al., 1994; Cassens et al., 2000; Hamilton et al., 2001). Milinkovitch et al. (1994) were the first to demonstrate, using 1352 bp of two mitochondrial ribosomal and cytochrome b genes, that Inia is a sister species of the superfamily Delphinoidea and within this superfamily, Delphinapterus leucas (Beluga) is the most closely related to Inia. In fact, the possible sister phylogenetic relationship between Inia and the Delphinoidea had been previously claimed based on morphometric studies (Heyning and Mead, 1990) and myoglobin data (McKenna, 1987). Also, Cassens et al. (2000), employing mitochondrial 12S and 16S rRNA and cyt-b genes as well as two nuclear genes (the gene encoding the interphotoreceptor retinoid-binding protein and lactalbumin gene), showed that Inia is the sister genus of another partially freshwater river dolphin (Pontoporia blainvillei) and both are the sister clade of Delphinoidea, with Monodontidae and Phocoenidae being the families least divergent from Inia. The divergence time of Inia and Pontoporia from Delphinoidea was around 31 millions of years ago, based on a maximum likelihood tree. The molecular trees of Arnason and Gullberg (1996) and Hamilton et al. (2001) also revealed that Iniidae is the sister clade of Delphinoidea, being less differentiated from Monodontidae and Phocoenidae than the Delphinidae, which radiated more recently. However, the phylogenetic position of the Baiji (Lipotes vexillifer), the Yangtze river dolphin, is questionable. Several studies from a morphological and paleontological perspective (Heyning, 1989; Barnes, 1990) and based on molecular evidence (Yang and Zhou, 1999) have postulated that Lipotes (Lipotidae) is the sister clade of Iniidae and Pontoporidae. Our results disagree with this view point, because Lipotes clustered in a different clade. Our Mhc sequences are more in agreement with the morphological studies of Muizon (1991) and Messenger and McGuire (1998), as well as with the molecular analyses of Cassens et al. (2000) and Hamilton et al. (2001), which showed that the real sister clade of Iniidae and Pontoporidae is Delphinoidea. Nonetheless, Mhc has a trans-specific mode of evolution; therefore, the genetic lineages usually are more ancient than the species split (Nei and Huges, 1991). Moreover, the selective patterns affecting the Mhc genes of Lipotes in the Chinese rivers could be quite different from the selective patterns regulating the Mhc genes in the Amazon and Orinoco basins. Another marked feature is the relevant amount of nucleotide diversity and genetic differentiation among the diverse Mhc cetacean sequences analyzed and within the genus Inia as well. Contrarily, there is an extremely conservative evolution of other molecular markers, such as autosome and Y chromosome introns in Inia geoffrensis and Inia boliviensis, where only six nucleotide substitutions were determined among 4544 bp analyzed (Ruiz-García et al., 2006a). Identically, DNA microsatellites seem to be extremely conserved in cetaceans; Schlötterer et al. (1991) estimated a rate of neutral nucleotide substitution of about 0.09% per million years, which is quite low. In an identical fashion, Ruiz-García et al. (2006b) estimated average microsatellite mutation rates for three cetaceans, Inia boliviensis, Pontoporia blainvillei and Sotalia fluviatilis, all of them living in South American freshwater basins. The estimates obtained ranged from 9.71 x 10-6 to 3.7 x 10-5, which were around the lower limit found in mammals. This means that some restrictive natural selection forces are acting upon the cetacean genomes. Contrarily, Mhc DQB-1 sequences seem to diverge more quickly than other nuclear sequences, supporting the possibility of different selective patterns affecting the MHC genes in these organisms, which could explain supposed inconsistencies in the cetacean phylogeny obtained by means of the MHC sequences. In conclusion, we present the first DQB-1 sequence relationships of this river dolphin genus with other cetaceans. Additionally, we provided evidence that these sequences are polymorphic within the Inia genus. The sequences obtained for Inia boliviensis and Inia geoffrensis were different but highly related. This could indicate that similar pathogen pressures act upon the animals of the different Amazon rivers. These data are not yet, however, conclusive; more results could give important insights into understanding the genetic split of the Bolivian species from the other pink river dolphin as well as the different selective pressures affecting the diverse boto populations sampled by us in Peru, Colombia, Ecuador, Brazil, and Venezuela. ACKNOWLEDGMENTS Economic resources to carry out this study were obtained through Colciencias (Grant 1203-09-11239; Filogeografía, estructura poblacional y diversidad genética en dos especies de delfines de río, Inia boliviensis e Inia geoffrensis, mediante el uso de marcadores moleculares) and the Fondo para la Acción Ambiental (120108-E0102141; Estructura y conservación genética de los delfines de rio, Inia y Sotalia, en las cuencas de la Amazonía y Orinoquía). Additional thanks go to Dr. Fernando Trujillo (Omacha Fundation), Hugo Gálvez (Iquitos, Perú), Mariana Escovar (La Paz, Bolivia), Ariel Rodríguez (Colombia), Esteban Payán (Colombia), Dr. Diana Alvarez (Colombia), Pablo Escobar-Armel (Colombia), Armando Castellanos (Quito, Ecuador), and Isaias and his sons (Requena, Perú). Many thanks go to the Bolivian, Peruvian, Ecuadorian Ministries of Environment to facilitate obtaining the CITES and capture permissions, and especially to the Colección Boliviana de Fauna (Dr. Julieta Vargas) in La Paz (Bolivia). Thanks also go to the Pontificia Universidad Javeriana (Colombia) and to the Universidad de Baja California Sur for respective economic assistance. REFERENCES Arnason U and Gullberg A (1996). Cytochrome b nucleotide sequences and the identification of five primary lineages of extant cetaceans. Mol. Biol. Evol. 13: 407-417. Banguera-Hinestroza E, Cardenas H, Ruiz-Garcia M, Marmontel M, et al. (2002). Molecular identification of evolutionarily significant units in the Amazon River dolphin Inia sp. (Cetacea: Iniidae). J. Hered. 93: 312-322. Barnes LG (1990). The fossil record and evolutionary relationship of the genus Tursiops. In: The bottlenose dolphin (Leatherwood S and Reeves R, eds.). Academic Press, San Diego, CA, USA, pp. 3-26. Bergström TF, Erlandsson R, Engkvist H, Josefsson A, et al. (1999). Phylogenetic history of hominoid DRB loci and alleles inferred from intron sequences. Immunol. Rev. 167: 351-365. Best RC and da Silva VMF (1989). Biology, status and conservation of Inia geoffrensis in the Amazon and Orinoco River basins. Occasional Paper of the IUCN Species Survival Commission SSC 3: 23-33. Bontrop RE, Otting N, de Groot NG and Doxiadis GG (1999). Major histocompatibility complex class II polymorphisms in primates. Immunol. Rev. 167: 339-350. Brown JH, Jardetzky T, Saper MA, Samraoui B, et al. (1988). A hypothetical model of the foreign antigen binding site of class II histocompatibility molecules. Nature 332: 845-850. Brown JH, Jardetzky TS, Gorga JC, Stern LJ, et al. (1993). Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364: 33-39. Carrington M, Nelson GW, Martin MP, Kissner T, et al. (1999). HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283: 1748-1752. Cassens I, Vicario S, Waddell VG, Balchowsky H, et al. (2000). Independent adaptation to riverine habitats allowed survival of ancient cetacean lineages. Proc. Natl. Acad. Sci. USA 97: 11343-11347. Da Silva VMF (1994). Aspects of the biology of the Amazonian dolphin genus Inia and Sotalia fluviatilis. Ph.D. dissertation, University of Cambridge, Cambridge, UK. Doherty PC and Zinkernagel RM (1975). H-2 compatibility requirement for T-cell-mediated lysis of target cells infected with lymphocytic choriomeningitis virus. Different cytotoxic T-cell specificities are associated with structures coded for in H-2K or H-2D. J. Exp. Med. 141: 1427-1436. Edwards SV and Hedrick PW (1998). Evolution and ecology of MHC molecules: from genomics to sexual selection. Trends Ecol. Evol. 13: 305-311. Evermann JF, Heeney JL, Roelke ME, McKeirnan AJ, et al. (1988). Biological and pathological consequences of feline infectious peritonitis virus infection in the cheetah. Arch. Virol. 102: 155-171. Flores-Ramírez S, Urban-Ramírez J and Miller RD (2000). Major histocompatibility complex class I loci from the gray whale (Eschrichtius robustus). J. Hered. 91: 279-282. Flores-Ramírez SJ, Miller RD and Urban-Ramírez J (2004). Major histocompatibility complex I polymorphism in a cetacean: the gray whale (Eschrichtius robustus). Mar. Mammal Sci. 20: 262-273. Garrigan D and Hedrick PW (2003). Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution Int. J. Org. Evolution 57: 1707-1722. Hambuch TM and Lacey EA (2002). Enhanced selection for MHC diversity in social tuco-tucos. Evolution Int. J. Org. Evolution 56: 841-845. Hamilton H, Caballero S, Collins AG and Brownell Jr RL (2001). Evolution of river dolphins. Proc. Biol. Sci. 268: 549-556. Heyning JE (1989). Comparative facial anatomy of beaked whales (Ziphiidae) and a systematic revision among the families of extant Odontoceti. Contrib. Sci. Nat. Hist. Mus. LA. 405: 1-64. Heyning JE and Mead JG (1990) Sensory abilities of cetaceans: Laboratory and field evidence (Thomas J and Kastelein R, eds.). Plenum Publishing Corporation, New York, NY, USA, pp. 67-79. Heyning JE and Lento GM (2002) The evolution of marine mammals. In: Marine mammal biology (Hoelzel AR, ed.). Chapter 2. Blackwell Publishing, Oxford, UK. Hill AV (1999). The immunogenetics of resistance to malaria. Proc. Assoc. Am. Physicians 111: 272-277. Hill AV, Allsopp CE, Kwiatkowski D, Anstey NM, et al. (1991). Common west African HLA antigens are associated with protection from severe malaria. Nature 352: 595-600. Hoelzel AR, Stephens JC and O’Brien SJ (1999). Molecular genetic diversity and evolution at the MHC DQB locus in four species of pinnipeds. Mol. Biol. Evol. 16: 611-618. Hughes AL and Nei M (1989). Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc. Natl. Acad. Sci USA 86: 958-962. Jepson A, Banya W, Sisay-Joof F, Hassan-King M, et al. (1997). Quantification of the relative contribution of major histocompatibility complex (MHC) and non-MHC genes to human immune responses to foreign antigens. Infect. Immun. 65: 872-876. Kimura M (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111-120. Klein J and Takahata N (1990). The major histocompatibility complex and the quest for origins. Immunol. Rev. 113: 5-25. Klein J, Satta Y, Huigin CO and Takahata N (1993). The molecular descent of the major histocompatibility complex. Ann. Rev. Immunol. 11: 269-295. Kumar S, Tamura K and Nei M (1993). MEGA: molecular evolutionary genetics analysis, version 1.01. Pennsylvania State University, University Park, PA, USA. Lukas D, Bradley BJ, Nsubuga AM, Doran-Sheehy D, et al. (2004). Major histocompatibility complex and microsatellite variation in two populations of wild gorillas. Mol. Ecol. 13: 3389-3402. Lyles AM and Dobson AP (1993). Infectious disease and intensive management: population dynamics, threatened hosts, and their parasites. J. Zoo Wildlife Med. 24: 315-326. Manning CJ, Wakeland EK and Potts WK (1992). Communal nesting patterns in mice implicate MHC genes in kin recognition. Nature 360: 581-583. Martins AR and da Silva V (2004a). River dolphins and flooded forest: seasonal habitat use and sexual segregation of botos (Inia geoffrensis) in an extreme Cetacean environment. J. Zool. 263: 295-305. Martins AR and da Silva V (2004b). Number, seasonal movements, and residency characteristics of river dolphins in an Amazonian flood plain lake system. Can. J. Zool. 82: 1307-1315. McKenna MC (1987). Molecules and morphology in evolution: conflict or compromise? Cambridge University Press, Cambridge, England. Messenger SL and McGuire JA (1998). Morphology, molecules, and the phylogenetics of cetaceans. Syst. Biol. 47: 90-124. Mikkos S, Roed K, Schmutz S and Andersson L (1999). Monomorphism and polymorphism at MHC DRB loci in domestic and wild ruminants. Immunol. Rev. 167: 169-178. Milinkovitch M and Thewissen JGM (1997). Even-toed fingerprints on whale ancestry. Nature 388: 622-623. Milinkovitch MC, Meyer A and Powell JR (1994). Phylogeny of all major groups of cetaceans based on DNA sequences from three mitochondrial genes. Mol. Biol. Evol. 11: 939-948. Muizon C (1991). A new Ziphiidae from the Early Miocene of Washington State and a phylogenetic analysis of the major groups of odontocetes. Bull. Mus. Natl. Hist. Nat. Paris 3-4: 279-326. Munguía-Vega A (2002). Estudio del complejo principal de histocompatibilidad en la historia evolutiva y demográfica de la vaquita Phocoena sinus. M.Sc. thesis, Centro de Investigaciones Biológicas del Noroeste, La Paz, México. Murray BW and White BN (1998). Sequence variation at the major histocompatibility complex DRB loci in beluga (Delphinapterus leucas) and narwhal (Monodon monoceros). Immunogenetics 48: 242-252. Murray BW, Malik S and White BN (1995). Sequence variation at the major histocompatibility complex locus DQ beta in beluga whales (Delphinapterus leucas). Mol. Biol. Evol. 12: 582-593. Murray DL, Kapke CA, Evermann JF and Fuller TK (1999). Infections disease and the conservation of free-ranging large carnivores. Anim. Conserv. 2: 241-254. Nei M and Hughes AL (1991) Polymorphism and evolution of the MHC in mammals. In: Evolution at the molecular level (Selander RK, Clark AG and Wittman TS, eds.). Sinauer Associates, Sunderland, MA, USA, pp. 222-247. Ober C, Elias S, Kostyu DD and Hauck WW (1992) Decreased fecundability in Hutterite couples sharing HLA-DR. Am. J. Hum. Genet. 50: 6-14. Potts WK and Wakeland EK (1993). Evolution of MHC genetic diversity: a tale of incest, pestilence and sexual preference. Trends Genet. 9: 408-412. Potts WK, Manning CJ and Wakeland EK (1991). Mating patterns in seminatural populations of mice influenced by MHC genotype. Nature 352: 619-621. Ruiz-García M, Caballero S and Martinez-Aguero M (2006a) Molecular differentiation among Inia geoffrensis and Inia boliviensis (Iniidae, Cetacea) by means of nuclear intron sequences. Genes Genet. Syst. (in press). Ruiz-García M, Escobar-Armel P, Caballero S and Secchi E (2006b). Determination of microsatellite mutation rates and effective numbers in three cetacean species (Inia boliviensis, Pontoporia blainvillei and Sotalia fluviatilis): comparisons with terrestrial mammals. Mol. Biol. Evol. (in press). Saitou N and Nei M (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406-425. Sambrook J, Fritsch EF and Maniatis T (1989). Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor Laboratoty Press, Cold Spring Harbor, NY, USA. Schlötterer C, Amos B and Tautz D (1991). Conservation of polymorphic simple sequence loci in cetacean species. Nature 354: 63-65. Slade RW (1992). Limited MHC polymorphism in the southern elephant seal: implications for MHC evolution and marine mammal population biology. Proc. Biol. Sci. 249: 163-171. Thompson JD, Higgins DG and Gibson TJ (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. Trowsdale J, Groves V and Arnason A (1989). Limited MHC polymorphism in whales. Immunogenetics 29: 19-24. Wedekind C, Seebeck T, Bettens F and Paepke AJ (1995). MHC dependent mate preferences in humans. Proc. Biol. Sci. 260: 245-249. Yang G and Zhou K (1999). A study on the molecular phylogeny of river dolphins. Acta Theriol. Sin. 19: 1-9. Yang G, Yan J, Zhou K and Wei F (2005). Sequence variation and gene duplication at MHC DQB loci of baiji (Lipotes vexillifer), a Chinese river dolphin. J. Hered. 96: 310-317. Yuhki N and O’Brien SJ (1990). DNA variation of the mammalian major histocompatibility complex reflects genomic diversity and population history. Proc. Natl. Acad. Sci. USA 87: 836-840. |
|