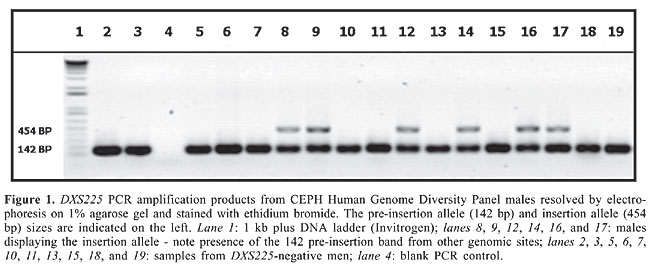
ABSTRACT. We describe a novel polymorphic Alu insertion (DXS225) on the human X chromosome (Xq21.3) embedded into an L1 retrotransposon. The DXS225 polymorphism was genotyped in 684 males from the CEPH Human Genome Diversity Panel. This insertion was found in all regions of the globe, suggesting that it took place before modern humans spread from Africa ca. 100,000 years ago. However, only one Amerindian population (Karitiana) showed this insertion allele, which may have been introduced by European admixture. Thus, it appears likely that the Alu insertion was absent from pre-Columbian America. Analysis of molecular variance worldwide demonstrated that 92.2% of the genetic variance was concentrated within populations. DXS225 is flanked by two microsatellites (DXS8114 and DXS1002), which are 86 kb apart and are in very strong linkage disequilibrium. The combination of a unique event polymorphism on the X chromosome in linkage disequilibrium with two rapidly evolving microsatellites should provide a useful tool for studies of human evolution. Key words: Alu, Polymorphism, X chromosome, CEPH panel, Human population genetics INTRODUCTION The human genome can be seen as a mosaic of discrete haplotype blocks, each with its own distinct genealogical history and variation pattern (reviewed by Paabo, 2003). Two of these haplotype blocks, the mitochondrial DNA and the non-recombining portion of the human Y chromosome, have been extremely useful in elucidating matrilineal and patrilineal genealogical patterns in human evolution, respectively (Cavalli-Sforza and Feldman, 2003). We reasoned that the identification of long haplotype blocks on the human X chromosome would provide another useful phylogeographical tool. With this in mind, we decided to study a region located between Xq13.3 and Xq21.3, with a recombination rate of 0.6 cM/Mb, a low rate when compared with the average X chromosome recombination rate of 1.3 cM/Mb (Nagaraja et al., 1997). Within this region we located five microsatellites (DXS995, DXS8076, DXS8114, DXS1002, and DXS1050) that had not shown recombination in 291 meioses tested during construction of the X chromosome genetic map (Dib et al., 1996). We analyzed the pairwise level of linkage disequilibrium between these microsatellites in an international panel of human DNA. No significant linkage disequilibrium between the most external loci, DXS995 and DXS1050, was observed (Pereira and Pena, 2006). Thus, even though recombination may be absent within short time spans, as seen in the CEPH pedigrees, on a long-term basis recombination and mutation did occur often enough to dissipate linkage disequilibrium. On the other hand, the microsatellites DXS1002 and DXS8114, which are 86 kb apart, remained in strong linkage disequilibrium (Pereira and Pena, 2006). We searched this 86-kb region for unique event polymorphisms, especially polymorphic Alu insertions. Screening for young Alu elements over the 86-kb region led to the identification of an AluYa5 sequence, embedded within a LINE-1 element, which proved to be polymorphic in humans. We surveyed the worldwide frequency distribution of this new polymorphic Alu insertion (named DXS225) in 684 males from the Human Genome Diversity Panel (Cann et al., 2002). MATERIAL AND METHODS DNA samples The DNA samples from unrelated Brazilians have been described previously (Alves-Silva et al., 2000). The HGDP-CEPH Diversity Panel (Cann et al., 2002) was provided by the Fondation Jean Dausset, Paris, France. This panel contains genetic material from 684 male individuals representing all five continents and belonging to 52 different populations from seven regional groups (Africa, Europe, Middle East, Central/South Asia, East Asia, Oceania, and America). A sample of male chimpanzee DNA was obtained from the Coriell Cell Repository (Camden, New Jersey). Additionally, we also typed 34 Amerindian samples that were kindly provided by Dr. Judith Kidd from Yale University, distributed as follows: 25 Ticuna (from Brazil) and 9 Muskoke (from the United States). Identification and testing of potential polymorphic Alu elements in a low recombination region of the X chromosome Using the Repeat Masker program (Smit et al., 2004), we searched an 86-kb region of human chromosome X (from position 8823714 to 8910624 on contig NT_011651.15) for Alu sequences from Ya5/8 families, which have been described as the most polymorphic Alu elements in the human genome (Batzer et al., 1995). One insertion from family Ya5 was identified, and primers were designed to amplify it as follows: AluYa5-F tccagaatctgcaaagaact, AluYa5-R- atgaccagtgatgaagagct. The primers were used to amplify pooled DNA samples from Brazilians. Each pool had 10 individuals. The PCR reactions were performed using a protocol that involved “hot start” and “step down” strategies (Hecker and Roux, 1996; Roux and Hecker, 1997). The hot start was implemented by a denaturing step of 94°C for 5 min, after which the Taq polymerase was added and then the reaction mix was subjected to 20 cycles of 94°C/30 s, 64°C/30 s (decreasing 1°C per cycle during the first five cycles) and 72°C/1 min. The PCR products were resolved by electrophoresis on 6% polyacrylamyde gels and silver stained (Santos et al., 1993) or on 1% agarose gels stained with ethidium bromide. Ascertaining the map position of polymorphic Alu To confirm the location of the polymorphic Ya5 sequence on the X chromosome identified using the above approach we amplified an X Chromosome Deletion Panel obtained from the Coriell Cell Repository (Camden, New Jersey). Statistical analysis The frequency of the polymorphic insertion was calculated by gene counting. The genetic structure was investigated by the analyses of molecular variance (AMOVA) (Excoffier et al., 1992) using the package Arlequin 2.0 (Schneider et al., 2000). RESULTS The identification and PCR amplification of Ya5 The 86-kb region studied was located in intron 1 of the DACH2 [Homo sapiens dachshund homolog 2 (Drosophila)] gene. The identified Alu sequence was flanked by identifiable short direct repeats, with a long poly-A tail at the 3’ end, and was inserted into a LINE-1 repetitive element. The Alu sequence was approximately 10 kb from DXS1002 and 76 kb from DXS8114. The primer pair flanking the Ya5 insertion amplified fragments corresponded to presence (insertion allele; 454 bp) and absence (pre-insertion allele; 142 bp) of the Alu element in the pooled samples from Brazil, Africa, Europe and Asia, while the pooled Native American samples and the chimpanzee DNA showed absence of the Ya5 insertion (data not shown). Amplification of individual human DNA samples confirmed the polymorphic nature of the Alu insertion, with a peculiarity: male samples showed either only the 142-bp product or both 142-bp and 454-bp products (Figure 1). A priori this is an unexpected result, as males are hemizygous for the X chromosome. However, since the Ya5 sequence is inserted into an L1 element, the primers that flank it most likely also amplify other homologous L1 sequences in the genome. This cross-reactivity makes it impossible to distinguish female insertion/insertion homozygotes and insertion/pre-insertion heterozygotes using conventional PCR. The distinction can be made using quantitative real-time PCR, if needed. However, for population studies, the typing of males is easy and straightforward and also allows direct establishment of haplotypes.
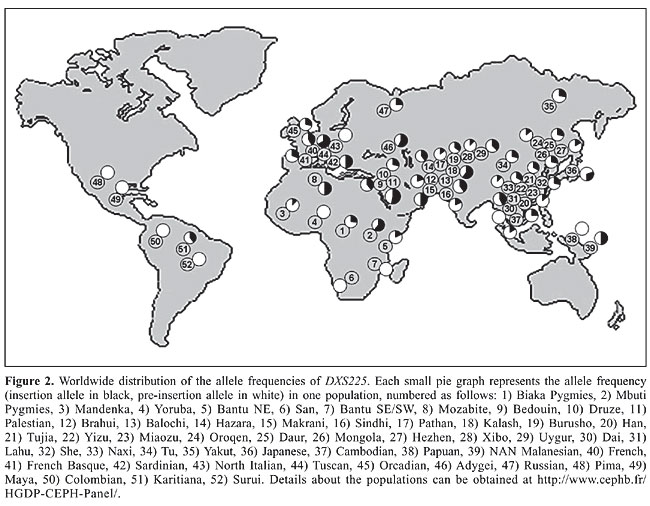
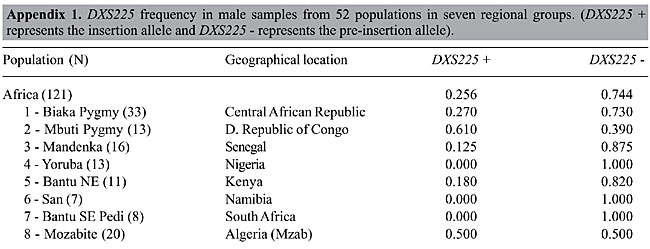
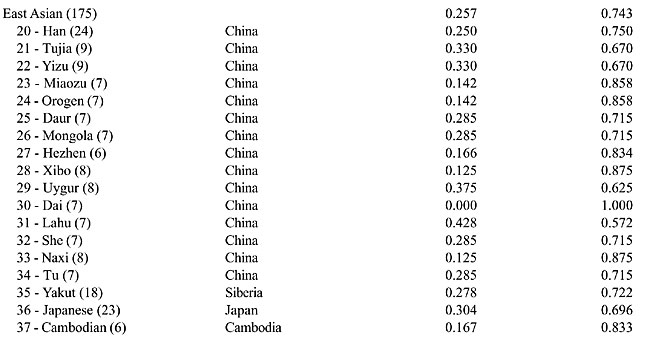
The PCR amplification of the X chromosome Ya5 sequence in the X chromosome deletion panel The deletion panel makes use of human-hamster or human-mouse somatic hybrid lines containing only the human X chromosome with deletions of different sizes. Since the hybrids originate from different individuals, there were lineages with and without the Ya5 Alu insertion. Differently from the typing of male human DNA, no amplification of the 142-bp product was seen in the insertion cases (data not shown). This means that the sequences co-amplifying the pre-insertion allele with the L1 primers are not located on the human X chromosome. As expected, the deletion panel mapped the Ya5 insertion in the Xq21 region deletion (data not shown). The novel X chromosome polymorphic Alu insertion was given the Genome Data Base access number 11524531 and received the name DXS225. DXS225 in males from the Human Genome Diversity project panel The DXS225 Alu insertion polymorphism was genotyped in 684 males from the CEPH Human Genome Diversity Panel. All regions of the globe showed the presence of the insertion, with frequencies of 0.256, 0.407, 0.347, 0.257, 0.190, 0.360, and 0.100 in Africa, Middle East, Central Asia, East Asia, Oceania, Europe, and America, respectively (Figure 2). The frequency of DXS225 insertion in each of the 52 populations of the panel is provided in Appendix 1. We subjected the DXS225 frequency data to AMOVA using the Arlequin software (Schneider et al., 2000) and found that 92.2% of the variability could be explained by differences among individuals within populations, 5.0% could be explained by differences among populations within regional groups and only 2.7% could be explained by differences among regional groups.
DXS225 in additional Amerindian samples We also typed the DXS225 polymorphism in 25 Ticuna and 9 Muskoke Amerindian males. All samples exhibited only the pre-insertion allele. DISCUSSION The human X chromosome presents features that make its study especially interesting for human evolutionary research: it is hemizygous in males, it has an effective population size that is intermediate between autosomes and Y chromosomes, and in every generation one third of its number switches sexes from males to females and one third from females to males (Schaffner, 2004). We describe a novel polymorphic Alu insertion (DXS225) on Xq21.3, embedded into an L1 retrotransposon. The polymorphic nature of the Alu insertion, DXS225, suggests that it happened in the human lineage after the divergence of hominids and the other primates. Indeed, we did not find the Alu insertion in an orthologous position in chimpanzee DNA. Moreover, the fact that the Alu sequence could be found in polymorphic frequencies all over the world allows the inference that this insertion took place before modern humans spread from Africa ca. 100,000 years ago. AMOVA showed that 92.2% of the genetic variance is concentrated within populations. This low level of genetic structure, plus the fact that the Alu insertion is embedded into an L1 repeat inside intron 1 of the DACH2 gene, suggest that DXS225 is most likely selectively neutral and that the variations in insertion allele frequencies among populations result from genetic drift. Among the five Amerindian populations studied, only the Karitiana showed presence of the Alu insertion. The Karitiana constitutes a very small group (estimated to have reached only 64 individuals in 1970) and in fact represents a single extended family (Storto and Velden, 2005). It is known that they had contact with European and African descendants in the early 20th century and it is thus conceivable that the Alu insertion allele was introduced into their gene pool by admixture. If that is the case, the pre-insertion allele may prove to have been monomorphic in pre-Columbian Amerindians, conceivably because of a founder effect. To test this notion, we tested 34 additional Amerindian samples (25 Ticuna from Brazil and 9 Muskoke from the United States) and in all of them we found only the pre-mutation allele. If further research confirms these findings, DXS225 may provide a useful marker for admixture studies. Recently, a comprehensive search for Alu insertion polymorphisms on the human X chromosome was undertaken and 16 such polymorphisms were identified (Callinan et al., 2003). However, these authors did not search within L1 retrotransposons, which are quite abundant on the X chromosome (Bailey et al., 2000) and that seems to be the reason why they did not find DXS225. This newly characterized Alu polymorphism is flanked by two microsatellites (DXS8114 and DXS1002), which are 86 kb apart and are in very strong linkage disequilibrium with each other. As shown by others (Tishkoff et al., 1996, 2000; Mountain et al., 2002; Gaspar et al., 2004; Ramakrishnan and Mountain, 2004) this combination of a unique event polymorphism on the X chromosome in linkage disequilibrium with two fast evolving microsatellites should provide a very useful tool for studies of human evolution. ACKNOWLEDGMENTS Research partially supported by a grant from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). R.W. Pereira received a PhD fellowship from the Conselho de Aperfeiçoamento do Pessoal de Ensino Superior (CAPES). We are grateful to Dr. Howard Cann of the Fondation Jean Dausset who provided the HGDP-CEPH Diversity Panel and to Dr. Judith Kidd from Yale University who made available Amerindian DNA samples. Neuza A. Rodrigues and Kátia Barroso provided expert technical assistance. REFERENCES Alves-Silva J, da Silva SM, Guimaraes PE, Ferreira AC et al. (2000). The ancestry of Brazilian mtDNA lineages. Am. J. Hum. Genet. 67: 444-461. Bailey JA, Carrel L, Chakravarti A and Eichler EE (2000). Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc. Natl. Acad. Sci. USA 97: 6634-6639. Batzer MA, Rubin CM, Hellmann-Blumberg U, Alegria-Hartman M et al. (1995). Dispersion and insertion polymorphism in two small subfamilies of recently amplified human Alu repeats. J. Mol. Biol. 247: 418-427. Callinan PA, Hedges DJ, Salem AH, Xing J et al. (2003). Comprehensive analysis of Alu-associated diversity on the human sex chromosomes. Gene 317: 103-110. Cann HM, de Toma C, Cazes L, Legrand MF et al. (2002). A human genome diversity cell line panel. Science 296: 261-262. Cavalli-Sforza LL and Feldman MW (2003). The application of molecular genetic approaches to the study of human evolution. Nat. Genet. (Suppl 33): 266-275. Dib C, Faure S, Fizames C, Samson D et al. (1996). A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380: 152-154. Excoffier L, Smouse PE and Quattro JM (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479-491. Gaspar P, Seixas S and Rocha J (2004). Genetic variation in a compound short tandem repeat/Alu haplotype system at the SB19.3 locus: properties and interpretation. Hum. Biol. 76: 277-287. Hecker KH and Roux KH (1996). High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. Biotechniques 20: 478-485. Mountain JL, Knight A, Jobin M, Gignoux C et al. (2002). SNPSTRs: empirically derived, rapidly typed, autosomal haplotypes for inference of population history and mutational processes. Genome Res. 12: 1766-1772. Nagaraja R, MacMillan S, Kere J, Jones C et al. (1997). X chromosome map at 75-kb STS resolution, revealing extremes of recombination and GC content. Genome Res. 7: 210-222. Paabo S (2003). The mosaic that is our genome. Nature 421: 409-412. Pereira RW and Pena SDJ (2006). Phylogeography of haplotypes of five microsatellites located in a low-recombination region of the X-chromosome: studies worldwide and in Brazilian populations. Genetica 126: 243-250. Ramakrishnan U and Mountain JL (2004). Precision and accuracy of divergence time estimates from STR and SNPSTR variation. Mol. Biol. Evol. 21: 1960-1971. Roux KH and Hecker KH (1997). One-step optimization using touchdown and stepdown PCR. Methods Mol. Biol. 67: 39-45. Santos FR, Pena SD and Epplen JT (1993). Genetic and population study of a Y-linked tetranucleotide repeat DNA polymorphism with a simple non-isotopic technique. Hum. Genet. 90: 655-656. Schaffner SF (2004). The X chromosome in population genetics. Nat. Rev. Genet. 5: 43-51. Schneider S, Roessli D and Excofier L (2000). Arlequin ver. 2000: a software for population genetics data analysis. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva, Switzerland. Smit AFA, Hubley R and Green P (2004). RepeatMasker Open-3.0. 1996-2004. [http://www.repeatmasker.org]. Storto L and Velden FFV (2005). Povos Indígenas do Brasil - Karitiana. [http://www.socioambiental.org/pib/epi/karitiana/karitiana.shtm]. Tishkoff SA, Dietzsch E, Speed W, Pakstis AJ et al. (1996). Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science 271: 1380-1387. Tishkoff SA, Pakstis AJ, Stoneking M, Kidd JR et al. (2000). Short tandem-repeat polymorphism/alu haplotype variation at the PLAT locus: implications for modern human origins. Am. J. Hum. Genet. 67: 901-925.
|
|