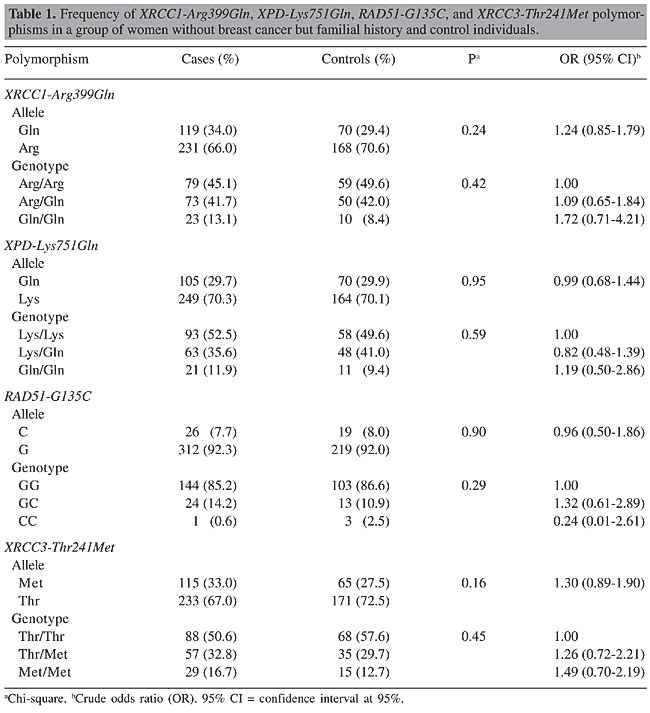
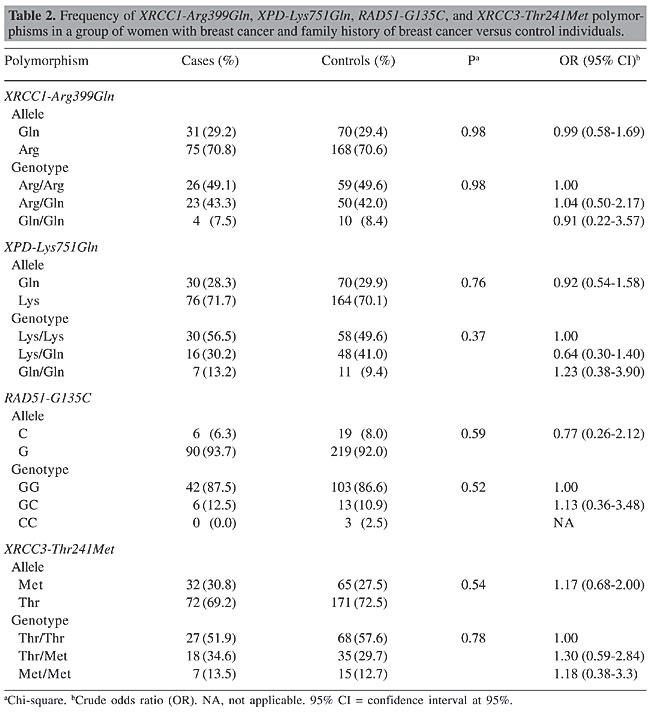
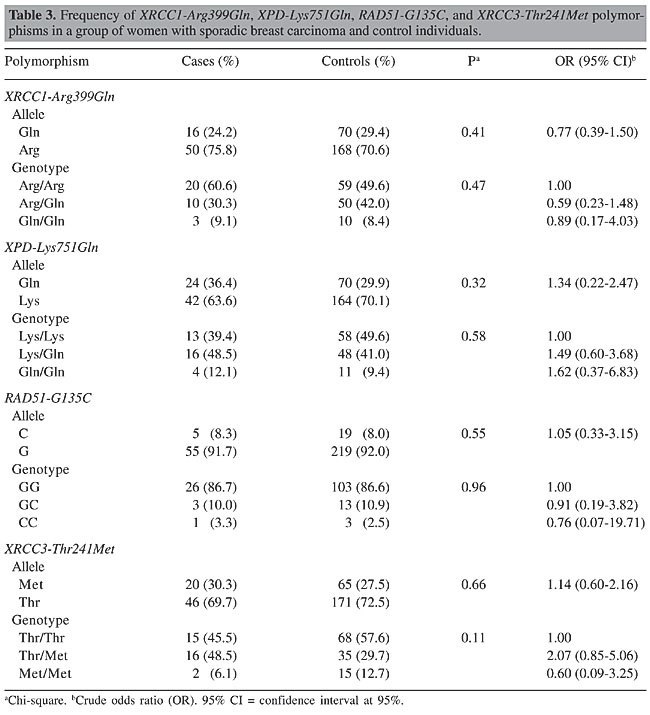
ABSTRACT. Several studies have reported that the genes involved in DNA repair and in the maintenance of genome integrity play a crucial role in protecting against mutations that lead to cancer. Epidemiologic evidence has shown that the inheritance of genetic variants at one or more loci results in a reduced DNA repair capacity and in an increased risk of cancer. Polymorphisms have been identified in several DNA repair genes, such as XRCC1, XPD, XRCC3, and RAD51, but the influence of specific genetic variants on repair phenotype and cancer risk has not yet been clarified. This was a case-control study design with three case groups: 53 women with breast cancer and family history; 33 women with sporadic breast cancer; 175 women with no breast cancer but with family history. The control group included 120 women with no breast cancer and no family history. The PCR-RFLP method was used to analyze the XRCC1-Arg399Gln, XPD-Lys751Gln, XRCC3-Thr241Met, and RAD51-G135C polymorphisms. No statistically significant differences were found between the case groups and the control group for any of the polymorphisms analyzed, and also between the breast cancer and family history group and the sporadic breast cancer group. Sample sizes of women with breast cancer, whether familial or sporadic, were insufficient to show any small true differences between the groups, but we have to consider that currently there is no clear consensus with respect to the association of these polymorphisms with breast cancer risk. Considering the data available, it can be conjectured that if there is any risk association between these single-nucleotide polymorphisms and breast cancer, this risk will probably be minimal. The greater the risk associated with cancer, the smaller the sample size required to demonstrate this association, and the data of different studies are usually, therefore, more concordant. Key words: Breast cancer, XRCC1-Arg399Gln, XPD-Lys751Gln, XRCC3-Thr241Met,RAD51-G135C, Polymorphisms INTRODUCTION Many environmental factors, such as radiation, diet and the use of endogenous or exogenous estrogens, have been associated with the risk of developing breast cancer. Recently, it has been suggested that polymorphic differences may lead to differences in susceptibility to cancer development. Several studies have reported that the genes involved in DNA repair and in the maintenance of genome integrity play a crucial role in providing protection against the mutations that lead to cancer (Bohr, 1995; Jiricny and Nystrom-Lahti, 2000). Epidemiologic evidence has shown that the inheritance of genetic variants at one or more loci results in reduced DNA repair capacity and an increase in the risk of cancer (Helzlsouer et al., 1996; Sturgis et al., 1999). Polymorphisms in several DNA repair genes have been identified, but the influence of specific genetic variants on phenotype repair and on the risk of developing cancer has not yet been clarified. BRCA1 and BRCA2 are two well-known breast cancer-susceptibility genes and their mutations account for most of the known hereditary carcinomas. BRCA proteins form complexes with other proteins involved in DNA repair such as RAD51. A missense mutation in RAD51 (Arg150Glu) has been described in Japanese patients with bilateral breast cancer (Kato et al., 2000). Further studies suggest that RAD51 (135 C/G) is a clinically significant modifier that raises breast cancer risk within the set of hereditary breast cancers (Levy-Lahad et al., 2001). XRCC3, XRCC1 and XPD belong to another group of genes that are involved in DNA repair and some polymorphisms in these genes are associated with increased breast cancer risk (Liu et al., 1998). The Thr241Met genotype substitution in XRCC3 is a non-conservative change that does not reside in the ATP-binding domains, which are the only functional domains that have been identified in the protein. This polymorphism is therefore likely to play a role in modifying the risk of breast cancer. The XRCC1 gene (X-ray repair cross-complementing gene) plays a role in the base excision repair pathway and has a BRCA1 C-terminal domain (BRCT) which is characteristic of proteins involved in cycle checkpoint functions and DNA damage. The base excision repair system is activated by ionizing radiation and by alkylating agents that cause DNA base damage and strand breaks. The Arg399Gln polymorphism resides in the C-terminal domain of XRCC1 within a relatively non-conserved region between BRCT domains. These results suggest that this DNA repair gene (XRCC1), a codon 399 genotype, may influence breast cancer risk, perhaps by modifying the effects of different forms of environmental exposure. The XPD gene is involved in the nucleotide-excision repair pathway. This protein repairs a wide range of structurally unrelated lesions, such as bulky adducts and thymidine dimers (Braithwaite et al., 1999). It has been reported that normal individuals with the XPD 751Gln variant form a higher number of DNA adducts than never-smoking individuals with the XPD 751Lys polymorphism (Matullo et al., 2001b). Therefore, these polymorphisms could also be involved in modifying susceptibility to carcinogenesis of the breast. Breast cancer is the principal cause of death from cancer in women in Brazil as well as in most of the more developed countries. Furthermore, to our knowledge, the prevalence of these polymorphic DNA repair genes has not yet been established in the Brazilian population. Moreover, the possible association of these genotypic variants with breast cancer susceptibility has not yet been fully evaluated. The aim of this study was to determine the frequency of XRCC1-Arg399Gln, XPD-Lys751Gln, XRCC3-Thr241Met, and RAD51-G135C in a sample of women in Campinas, Brazil, and to evaluate their association with breast cancer susceptibility, using a case-control study. MATERIAL AND METHODS Patient selection Women were enrolled into four groups as follows: 53 women with breast cancer and family history of breast cancer; 33 women with sporadic breast carcinoma; 175 women with no breast cancer but with family history, and 120 women with no breast cancer and no family history. The last group served as the control group for this study. The women with breast cancer who were included in this study were receiving care at the Centro de Atenção Integral à Saúde da Mulher (CAISM), UNICAMP, Brazil. The criteria for inviting the women with a family history of breast cancer to participate in the study were: early onset (less than 35 years of age); bilateral carcinoma; more than three cases of breast cancer and more than one case of ovarian cancer in the family; more than two first-degree relatives involved, and male breast cancer. The women who did not have breast cancer were selected from volunteers among hospital personnel in the region of Campinas, and were classified according to family history of breast cancer. Those with more than three cases of breast cancer and more than one case of ovarian cancer in the family, or who had one or more first-degree relatives with breast cancer, or who had a case of male breast cancer in the family, were considered to have a positive family history. The control group was made up of women who had no known cases of breast cancer in any relative, first-degree or otherwise. PCR-RFLP analysis DNA was isolated from peripheral leukocytes obtained from the women. Polymerase chain reaction (PCR) followed by enzymatic digestion (RFLP) was used for genotyping the XRCC1-Arg399Gln, XPD-Lys751Gln, XRCC3-Thr241Met, and RAD51-G135C polymorphisms. All the PCR reactions were carried out in a total reaction volume of 50 µL containing nearly 100 ng genomic DNA, 1 U Taq polymerase in 1X PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, and 0.20 µM of each primer. Thermal cycling conditions were as follows: initial denaturation step at 95°C for 3 min, 35 cycles of PCR consisting of 95°C for 30 s, 60°, 55°, 60°, and 53°C for 30 s for XPD, XRCC1, XRCC3, and RAD51 genes, respectively, and 72°C for 30 s, followed by a final extension step at 72°C for 10 min. The XPD-Lys751Gln polymorphism was determined using the following primers: sense, 5’-CTGCTCAGCCTGGAGCAGCTAGAATCAGAGGAGACGCTG-3’; anti-sense, 5’-AAGACCTTCTAGCACCACCG-3’, resulting in a 161-bp PCR product. This was digested with PstI restriction enzyme. The digestion resulted in 41- and 120-bp fragments corresponding to the Gln751 allelic variant or a 161-bp fragment containing the Lys751 allele. The XRCC1-Arg399Gln polymorphism was determined using the following primers: sense, 5’-CAAGTACAGCCAGGTCCTAG-3’; antisense, 5’-CCTTCCCTCATCTGGAGTAC-3’. The 248-bp PCR product was digested with NciI restriction enzyme. The Arg399 allele was represented by fragments of 89 and 159 bp, and the Gln399 allele (variant allele) was not digested. The XRCC3-Thr241Met polymorphism was determined using the following primers: sense, 5’-GCCTGGTGGTCATCGACTC-3’; anti-sense, 5’-ACAGGGCTCTGGAAGGCACTGCTCAGCTCACGCACC-3’, resulting in a 136-bp PCR product. This was digested with NcoI restriction enzyme. The Thr241 allele was represented by 39- and 97-bp fragments, and the Met241 allele (variant allele) was not digested. The RAD51-G135C polymorphism was determined using the following primers: sense, 5’-TGGGAACTGCAACTCATCTGG-3’; anti-sense, 5’-GCGCTCCTCTCTCCAGCA-3’, resulting in a 157-bp PCR product. This was digested with MvaI restriction enzyme. The digestion resulted in 86- and 71-bp fragments corresponding to the G135 allele, or a 161-bp fragment representing the C135 allele (variant allele). The PCR products were visualized by electrophoresis on 2% agarose gel, and the digestion products were visualized by electrophoresis on 3% agarose gel. PCR followed by enzymatic digestion was performed for genotyping the XRCC1-Arg399Gln, RAD51-G135C, XPD-Lys751Gln, and XRCC3-Thr241Met polymorphisms. Statistical analysis Chi-square analysis (c2 tests) was used to test the association between the genotypes and alleles in relation to the cases and controls. P < 0.05 was used as the criterion of significance. The odds ratio (OR) and their 95% confidence intervals (CI) were calculated to estimate the strength of the association between polymorphism genotype alleles and cases and controls. RESULTS No statistically significant differences were observed in the alleles or in the genotype frequencies of the XRCC1-Arg399Gln, XPD-Lys751Gln, XRCC3-Thr241Met, and RAD51-G135C gene polymorphisms between the control group and the women with breast cancer and family history of breast cancer (Table 1), between the control group and the women with sporadic breast cancer (Table 2), nor between the control group and the women with no breast cancer but with a family history of breast cancer (Table 3).
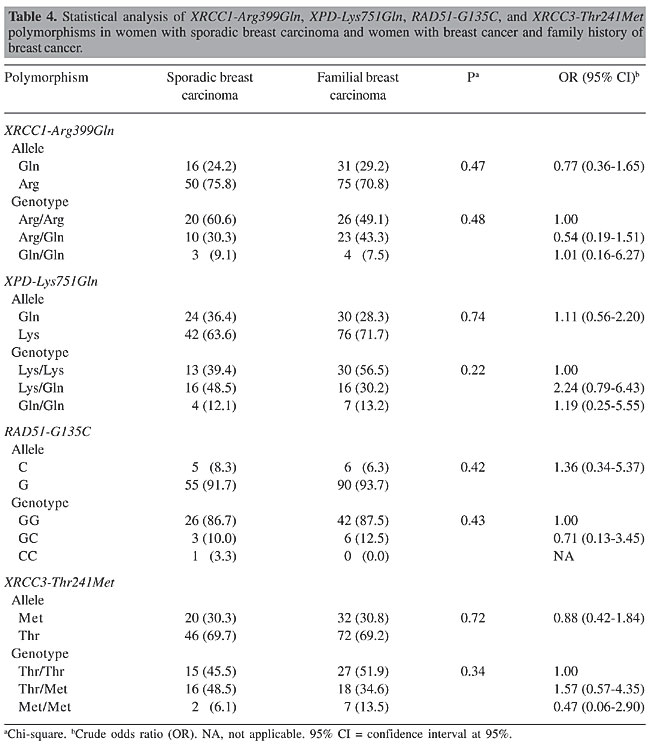
The women with sporadic breast cancer showed an incidence of 45.5, 48.5 and 6.1%, respectively, for the Thr/Thr, Thr/Met, and Met/Met genotypes of the XRCC3 gene, whereas the control group showed 57.6, 29.7, and 12.7% for the same genotypes. The Thr/Met genotype frequency came close to statistical significance with an OR of 2.07 and 95% CI of 0.85-5.06, considering the Thr/Thr genotype as reference (Table 3). Because we were interested in the association between the genotype found in cases of sporadic cancer and the genotype identified in cases of women with a family history of breast cancer, these data were also analyzed. However, we found no statistically significant difference in the variant allele frequencies of these DNA repair genotypes (Table 4).
DISCUSSION The present study examined whether polymorphisms in four DNA repair genes involved in base excision, nucleotide excision, and homologous double-stranded DNA repair pathways are related to the development of familial breast cancer. We found no association between familial breast cancer and the XRCC1-Arg399Gln, XPD-Lys751Gln, XRCC3-Thr241Met, and RAD51-G135C polymorphisms in this study population. Evidence suggests that the difference in DNA repair capacity among individuals is genetically determined. The phenotype of reduced repair capacity for one pathway is independent from the phenotype for any other pathway (Chu and Mayne, 1996), which is consistent with the hypothesis that DNA repair is genetically regulated. Measurement of repair capacity in twins (Pero et al., 1983) and the elevated frequency of individuals with reduced repair capacity among relatives of cancer patients provide further evidence that repair capacity is a genetic trait (Kovacs and Almendral, 1987; Pero et al.,1989). This variation in DNA repair capacity has characteristics that would be expected from cancer susceptibility genes since it may be the reduced function of the proteins encoded by these alleles rather than the absence of this function that causes disease. These proteins exist at polymorphic frequency in the general population, and they exhibit incomplete penetrance (Mohrenweiser and Jones, 1998; Shen et al., 1998). The XRCC1-Arg399Gln gene polymorphism has been studied as a risk factor for various cancers. The variant Gln allele has been linked to an increased risk of lung cancer (Divine et al., 2001; Zhou et al., 2003), head and neck cancer (Sturgis et al., 1999) and possibly stomach cancer (Shen et al., 2000). On the other hand, this allele was reported to be associated with a reduced risk of bladder cancer (Stern et al., 2001), esophageal cancer (Lee et al., 2002) and non-melanoma skin cancer (Nelson et al., 2002). Null association was also reported for lung cancer (Cavalieri et al., 2000; Nelson et al., 2002). In relation to breast cancer, Duell et al. (2002) reported a positive association between breast cancer and XRCC1 codon 399 Arg/Gln or Gln/Gln genotypes compared with Arg/Arg among African Americans but not in white American women. Shu et al. (2003) showed that the XRCC1-Arg399Gln gene polymorphism alone did not appear to play a substantial role in the risk of breast cancer among Chinese women. Smith et al. (2003) found no association between the XRCC1-399 Gln/Gln genotype and breast cancer. However, other studies showed an increased risk of breast cancer with this polymorphism (Sigurdson et al., 2004; Figueiredo et al., 2004; Han et al., 2004a,b). The XRCC3 gene is involved in the homologous recombinational pathway of the DNA double-strand break repair and interacts directly with RAD51 (Liu et al., 1998; Johnson and Jasin, 2001). No association was found between this polymorphism and lung cancer (Sugimura et al., 1999; Xing et al., 2001), squamous cell carcinoma of the head and neck (Wikman et al., 2000; Benhamou et al., 2004), gastric cancer (Shen et al., 2004) or basal cell carcinoma (Jacobsen et al., 2003). A positive association that achieved statistical significance was shown between the XRCC3 gene and melanoma and bladder cancer (Winsey et al., 2000; Matullo et al., 2001a); however, these results were not confirmed in subsequent, larger studies (Duan et al., 2002; Stern et al., 2002). In relation to XRCC3-T241m, Han et al. (2004a) observed no significant elevation in the risk for breast cancer. There was some evidence of a combined effect of body mass index and this polymorphism on risk estimates, which led the investigators to suggest that this polymorphism may influence breast cancer risk by modifying the effect of risk factors such as family history (Figueiredo et al., 2004). Smith et al. (2003) provided evidence that a variant of the XRCC3 gene, particularly when found in combination with other variants, contributes to breast cancer susceptibility. Our study on genotype Thr/Met of XRCC3 gene polymorphisms showed results close to statistical significance with an OR of 2.07 and 95% CI of 0.85-5.06, considering the Thr/Thr genotype as a reference. Interestingly, this result was observed in women with sporadic breast cancer but not in those with breast cancer and family history. The study group of women with sporadic breast cancer included 33 patients and this sample may be statistically underpowered to show true differences or to reject any difference among the groups compared. XPD-L751G is the most commonly studied polymorphism of this gene, and no statistically significant findings have ever been reported with respect to any increased risk of bladder cancer (Matullo et al., 2001a), skin basal cell cancer (Vogel et al., 2001), non-small cell lung cancer (Butkiewicz et al., 2001), or melanoma (Winsey et al., 2000). This polymorphism appears to be connected with smoking status and may increase cancer risk among non-smokers (Zhou et al., 2002). In relation to breast cancer, subjects with the Gln/Gln genotype at codon 751 were found to have higher adduct levels in tumor tissue than in tissue from benign breast disease controls (Tang et al., 2002). Forsti et al. (2004) searched for low-penetrant genes by measuring the frequencies of single-nucleotide polymorphisms for the following genes: NBS1, XPC, XPD, XRCC1, XRCC3, MTHFR, and cyclin D1. They concluded that none of the polymorphisms tested were associated with breast cancer, with the probable exception of XPD. In fact, there is little data on the association between breast cancer risk and XPD polymorphism. The product of the RAD51 gene works in conjunction with BRCA1 and BRCA2 in the repair of double-stranded DNA breaks. Jakubowska et al. (2003) suggested that RAD51 may be a genetic modifier of breast cancer risk in BRCA1 carriers in the Polish population. Kadouri et al. (2004) showed that in non-carrier breast cancer cases, bearing RAD51-G135C was not associated with breast cancer risk, but they suggested that the risk may be significantly elevated in carriers of BRCA2 mutations who also carry a RAD51-135C allele. In BRCA1 carriers and non-carriers, no effect of this single-nucleotide polymorphism was found. Blasiak et al. (2003) suggested that the G/C polymorphism of the RAD51 gene may not be directly involved in the development and/or progression of breast cancer; therefore, it may not be useful as an independent marker of this disease. The functional significance of the single-nucleotide polymorphism of the DNA-repair gene in DNA repair and human breast cancer risk is currently the subject of intense study, and there are many challenges that must be met. The results available should be interpreted with caution and other more conclusive studies should be carried out. The discrepancies between the results currently available could be due to different subject sample sizes and different study designs. Diverse environmental exposure could also contribute to divergent results. The results of this study do not provide insights into the function of these polymorphisms at the cellular level, but they do indicate that these DNA-repair gene polymorphisms are not significantly associated with familial breast cancer in the study population. The sample sizes of the groups of women with breast cancer, women with familial breast cancer and women with sporadic breast cancer were not sufficiently large to detect any true differences between the groups, but we have to consider that currently there is no clear consensus with respect to the association of these polymorphisms with breast cancer risk. Considering the data available, it can be conjectured that if there is any risk association between these single-nucleotide polymorphisms and breast cancer, this risk will probably be minimal. The greater the risk associated with cancer, the smaller the sample size that would be required to demonstrate this association, and the data of different studies are usually, therefore, more concordant. ACKNOWLEDGMENTS The authors wish to thank the staffs of the involved institutions and all of the participants and volunteers for their contribution. Research supported by CAPES-Ministry of Education, Brazil, grant number BEX2448/02-5 and FLAD-Luso-Brazilian Development Foundation, grant number L-V-172/2002. REFERENCES Benhamou S, Tuimala J, Bouchardy C, Dayer P et al. (2004). DNA repair gene XRCC2 and XRCC3 polymorphisms and susceptibility to cancers of the upper aerodigestive tract. Int. J. Cancer 112: 901-904. Blasiak J, Przybylowska K, Czechowska A, Zadrozny M et al. (2003). Analysis of the G/C polymorphism in the 5'-untranslated region of the RAD51 gene in breast cancer. Acta Biochim. Pol. 50: 249-253. Bohr VA (1995). DNA repair fine structure and its relations to genomic instability. Carcinogenesis 16: 2885-2892. Braithwaite E, Wu X and Wang Z (1999). Repair of DNA lesions: mechanisms and relative repair efficiencies. Mutat. Res. 424: 207-219. Butkiewicz D, Rusin M, Enewold L, Shields PG et al. (2001). Genetic polymorphisms in DNA repair genes and risk of lung cancer. Carcinogenesis 22: 593-597. Cavalieri E, Frenkel K, Liehr JG, Rogan E et al. (2000). Estrogens as endogenous genotoxic agents: DNA adducts and mutations. J. Natl. Cancer Int. Monogr. 27: 75-93. Chu G and Mayne L (1996). Xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy: do the genes explain the diseases? Trends Genet. 12: 187-192. Divine KK, Gilliland FD, Crowell RE, Stidley CA et al. (2001). The XRCC1 399 glutamine allele is a risk factor for adenocarcinoma of the lung. Mutat. Res. 461: 273-278. Duan Z, Shen H, Lee JE, Gershenwald JE et al. (2002). DNA repair gene XRCC3 241Met variant is not associated with risk of cutaneous malignant melanoma. Cancer Epidemiol. Biomarkers Prev. 11: 1142-1143. Duell EJ, Holly EA, Bracci PM, Wiencke JK et al. (2002). A population-based study of the Arg399Gln polymorphism in X-ray repair cross complementing group 1 (XRCC1) and risk of pancreatic adenocarcinoma. Cancer Res. 62: 4630-4636. Figueiredo JC, Knight JA, Briollais L, Andrulis IL et al. (2004). Polymorphisms XRCC1-R399Q and XRCC3-T241M and the risk of breast cancer at the Ontario site of the Breast Cancer Family Registry. Cancer Epidemiol. Biomarkers Prev. 13: 583-591. Forsti A, Angelini S, Festa F, Sanyal S et al. (2004). Single nucleotide polymorphisms in breast cancer. Oncol. Rep. 11: 917-922. Han J, Hankinson SE, Ranu H, De Vivo I et al. (2004a). Polymorphisms in DNA double-strand break repair genes and breast cancer risk in the Nurses’ Health Study. Carcinogenesis 25: 189-195. Han J, Hankinson SE, Zhang SM, De Vivo I et al. (2004b). Interaction between genetic variations in DNA repair genes and plasma folate on breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 13: 520-524. Helzlsouer KJ, Harris EL, Parshad R, Perry HR et al. (1996). DNA repair proficiency: potential susceptibility factor for breast cancer. J. Natl. Cancer Inst. 88: 754-755. Jacobsen NR, Nexo BA, Olsen A, Overvad K et al. (2003). No association between the DNA repair gene XRCC3 T241M polymorphism and risk of skin cancer and breast cancer. Cancer Epidemiol. Biomarkers Prev. 12: 584-585. Jakubowska A, Narod SA, Goldgar DE, Mierzejewski M et al. (2003). Breast cancer risk reduction associated with the RAD51 polymorphism among carriers of the BRCA1 5382insC mutation in Poland. Cancer Epidemiol. Biomarkers Prev. 12: 457-459. Jiricny J and Nystrom-Lahti M (2000). Mismatch repair defects in cancer. Curr. Opin. Genet. Dev. 10: 157-161. Johnson RD and Jasin M (2001). Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 29: 196-201. Kadouri L, Kote-Jarai Z, Hubert A, Durocher F et al. (2004). A single-nucleotide polymorphism in the RAD51 gene modifies breast cancer risk in BRCA2 carriers, but not in BRCA1 carriers or noncarriers. Br. J. Cancer 90: 2002-2005. Kato M, Yano K, Matsuo F, Saito H et al. (2000). Identification of Rad51 alteration in patients with bilateral breast cancer. J. Hum. Genet. 45: 133-137. Kovacs E and Almendral A (1987). Reduced DNA repair synthesis in healthy women having first degree relatives with breast cancer. Eur. J. Cancer Clin. Oncol. 23: 1051-1057. Lee SG, Kim B, Choi J, Kim C et al. (2002). Genetic polymorphisms of XRCC1 and risk of gastric cancer. Cancer Lett. 187: 53-60. Levy-Lahad E, Lahad A, Eisenberg S, Dagan E et al. (2001). A single nucleotide polymorphism in the RAD51 gene modifies cancer risk in BRCA2 but not BRCA1 carriers. Proc. Natl. Acad. Sci. USA 98: 3232-3236. Liu N, Lamerdin JE, Tebbs RS, Schild D et al. (1998). XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell 1: 783-793. Matullo G, Guarrera S, Carturan S, Peluso M et al. (2001a). DNA repair gene polymorphisms, bulky DNA adducts in white blood cells and bladder cancer in a case-control study. Int. J. Cancer 92: 562-567. Matullo G, Palli D, Peluso M, Guarrera S et al. (2001b). XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis 22: 1437-1445. Mohrenweiser HW and Jones IM (1998). Variation in DNA repair is a factor in cancer susceptibility: a paradigm for the promises and perils of individual and population risk estimation? Mutat. Res. 400: 15-24. Nelson HH, Kelsey KT, Mott LA and Karagas MR (2002). The XRCC1 Arg399Gln polymorphism, sunburn, and non-melanoma skin cancer: evidence of gene-environment interaction. Cancer Res. 62: 152-155. Pero RW, Bryngelsson C, Bryngelsson T and Norden A (1983). A genetic component of the variance of N-acetoxy-2-acetylaminofluorene-induced DNA damage in mononuclear leukocytes determined by a twin study. Hum. Genet. 65: 181-184. Pero RW, Johnson DB, Markowitz M, Doyle G et al. (1989). DNA repair synthesis in individuals with and without a family history of cancer. Carcinogenesis 10: 693-697. Shen H, Xu Y, Qian Y, Yu R et al. (2000). Polymorphisms of the DNA repair gene XRCC1 and risk of gastric cancer in a Chinese population. Int. J. Cancer 88: 601-606. Shen H, Wang X, Hu Z, Zhang Z et al. (2004). Polymorphisms of DNA repair gene XRCC3 Thr241Met and risk of gastric cancer in a Chinese population. Cancer Lett. 206: 51-58. Shen MR, Jones IM and Mohrenweiser H (1998). Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 58: 604-608. Shu XO, Cai Q, Gao YT, Wen W et al. (2003). A population-based case-control study of the Arg399Gln polymorphism in DNA repair gene XRCC1 and risk of breast cancer. Cancer Epidemiol. Biomarkers Prev. 12: 1462-1467. Sigurdson AJ, Hauptmann M, Chatterjee N, Alexander BH et al. (2004). Kin-cohort estimates for familial breast cancer risk in relation to variants in DNA base excision repair, BRCA1 interacting and growth factor genes. BMC Cancer 12: 4-9. Smith TR, Miller MS, Lohman K, Lange EM et al. (2003). Polymorphisms of XRCC1 and XRCC3 genes and susceptibility to breast cancer. Cancer Lett. 190: 183-190. Stern MC, Umbach DM, van Gils CH, Lunn RM et al. (2001). DNA repair gene XRCC1 polymorphisms, smoking, and bladder cancer risk. Cancer Epidemiol. Biomarkers Prev. 10: 125-131. Stern MC, Umbach DM, Lunn RM and Taylor JA (2002). DNA repair gene XRCC3 codon 241 polymorphism, its interaction with smoking and XRCC1 polymorphisms, and bladder cancer risk. Cancer Epidemiol. Biomarkers Prev. 11: 939-943. Sturgis EM, Castillo EJ, Li L, Zheng R et al. (1999). Polymorphisms of DNA repair gene XRCC1 in squamous cell carcinoma of the head and neck. Carcinogenesis 20: 2125-2129. Sugimura H, Kohno T, Wakai K, Nagura K et al. (1999). hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Cancer Epidemiol. Biomarkers Prev. 8: 669-674. Tang D, Cho S, Rundle A, Chen S et al. (2002). Polymorphisms in the DNA repair enzyme XPD are associated with increased levels of PAH-DNA adducts in a case-control study of breast cancer. Breast Cancer Res. Treat. 75: 159-166. Vogel U, Hedayati M, Dybdahl M, Grossman L et al. (2001). Polymorphisms of the DNA repair gene XPD: correlations with risk of basal cell carcinoma revisited. Carcinogenesis 22: 899-904. Xing DY, Tan W, Song N and Lin DX (2001). Ser326Cys polymorphism in hOGG1 gene and risk of esophageal cancer in Chinese population. Int. J. Cancer 95: 140-143. Wikman H, Risch A, Klimek F, Schmezer P et al. (2000). hOGG1 polymorphism and loss of heterozygosity (LOH): significance for lung cancer susceptibility in a Caucasian population. Int. J. Cancer 88: 932-937. Winsey SL, Haldar NA, Marsh HP, Bunce M et al. (2000). A variant within the DNA repair gene XRCC3 is associated with the development of melanoma skin cancer. Cancer Res. 60: 5612-5616. Zhou W, Liu G, Miller DP, Thurston SW et al. (2002). Gene-environment interaction for the ERCC2 polymorphisms and cumulative cigarette smoking exposure in lung cancer. Cancer Res. 62: 1377-1381. Zhou W, Liu G, Miller DP, Thurston SW et al. (2003). Polymorphisms in the DNA repair genes XRCC1 and ERCC2, smoking, and lung cancer risk. Cancer Epidemiol. Biomarkers Prev. 12: 359-365. |
|