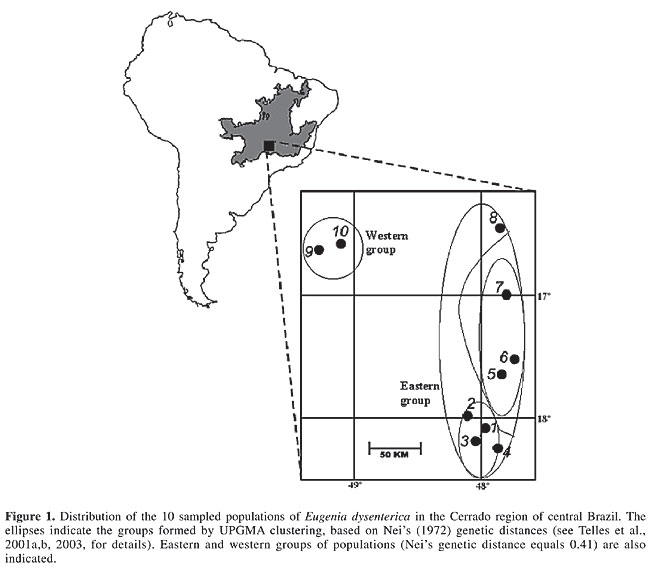
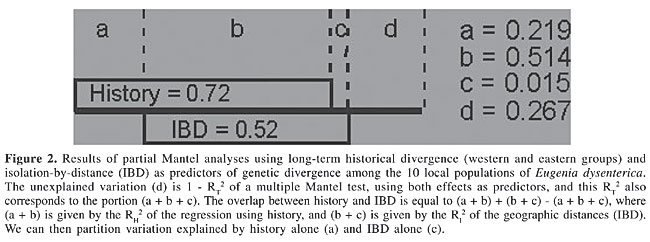
ABSTRACT. Mantel tests of matrix correspondence have been widely used in population genetics to examine microevolutionary processes, such as isolation-by-distance (IBD). We used partial and multiple Mantel tests to simultaneously test long-term historical effects and current divergence and equilibrium processes, such as IBD. We used these procedures to calculate genetic divergence among Eugenia dysenterica (Myrtaceae) populations in Central Brazil. The Nei’s genetic distances between pairs of local populations were strongly correlated with geographic distances, suggesting an IBD process, but field observations and the geographic distribution of the samples suggest that populations may have been subjected to more complex evolutionary processes of genetic divergence. Partial Mantel regression was used to partition the effects of geographic structure and long-term divergence associated with a possible historical barrier. The R2 of the model with both effects was 73.3%, and after the partition 21.9% of the variation in the genetic distances could be attributed to long-term historical divergence alone, whereas only 1.5% of the variation in genetic distances could be attributed to IBD. As expected, there was a large overlap between these processes when explaining genetic divergence, so it was not possible to entirely partition divergence between historical and contemporary processes. Key words: Mantel test, Isolation-by-distance, Isozymes, Eugenia, Genetic distances, Partial Mantel test INTRODUCTION Mantel test is a permutational procedure used to test the statistical significance of matrix correlations (Sokal, 1979; Manly, 1985, 1997; Epperson, 2003), and they have been widely used in population genetics to compare two or more distance matrices, testing hypotheses about spatial, temporal or environmental structures in genetic distances (Smouse et al., 1986; Diniz-Filho, 2000). In a more specific context, high correlations between genetic and geographic distances in continuous space have been primarily interpreted as resulting from isolation-by-distance (IBD) processes (see Congdon et al., 2000; Maes and Volckaert, 2002; Manel et al., 2003); but see Santos et al. (2003). However, before interpreting results of these matrix correlations, it is important to consider a basic problem in statistical analyses, i.e., the idea of achieving cause-effect relationships from correlations, based on non-experimental datasets obtained from nature (see Sokal and Rohlf, 1995; Shipley, 2002). In this case, a very obvious problem in (matrix) correlation analyses is the confounding effects of other variables, which could be in principle solved by using more complex multiple regression and correlation analyses. The use of these analyses begins by assuming that other predictor variables (or matrices) are known and could be measured in the same samples (see Telles et al., 2001a,b, for a recent example). Despite a few recent criticisms due to high type I error rates (see Raufaste and Rousset, 2001; Castellano and Balletto, 2002; Rousset, 2002), partial and multiple Mantel tests (Smouse et al., 1986; Legendre and Legendre, 1998) could be used to decompose the relative importance of these several factors involved in the structuring genetic distances, as recently done by Lampert et al. (2003). More specifically, when interpreting the matrix correlation between genetic and geographic distances, which are involved in IBD and related microevolutionary processes, a well-known effect that could confound this evaluation is the long-term historical divergence between two or more groups of populations (clades) in the study area (see Telles and Diniz-Filho, 2000; Epperson, 2003; Lampert et al., 2003). If this long-term divergence among clades is also structured in geographic space (as it currently is due to geographical barriers to dispersion and spatially structured colonization events), then a spurious interpretation of IBD under a simple Mantel test is possible. Although IBD creates a strong correlation between genetic divergence and geographic distance, this occurs due to a contemporary balance between local genetic drift and geographically mediated gene flow. We believe that, in such a situation, both phenomena (long-term historical divergence and current IBD) can be interpreted as confounding scaling effects in observed genetic distances and, under some circumstances, could be evaluated by multiple and partial Mantel tests. PARTITIONING LONG-TERM HISTORICAL DIVERGENCE AND ISOLATION-BY-DISTANCE If groups of local populations could be explicitly defined to diverge under long-term historical processes, multiple Mantel tests could be used to partition the contemporary (IBD) and historical effects. Long-term divergence can be inferred by “external” information (biogeographical and ecological data) or can be derived from phylogenetic analysis of certain specific genes (mtDNA markers, for example, in a phylogeographic context - Avise, 2000; see Santos et al., 2003, for a recent example). The groups of populations belonging to the same clade, or group, could be linked in a pairwise model matrix (see Manly, 1985, 1997; Sokal et al., 1997; Rodrigues et al., 2002), in which the value 1.0 indicates that two populations are “linked” (within the same group), and zero elsewhere. So, there are three different matrices to be analyzed: 1) genetic distances (obtained by many available methods, which vary by using different kinds of molecular markers - see Swofford et al., 1996); 2) geographic distances, and 3) a model matrix expressing long-term historical divergence. There are different ways to evaluate the relationship between these three matrices. The general idea is that the IBD process occurs on a much smaller time-scale than long-term historical isolation or deep-time coalescence. So, the obvious and simpler solution would be to apply Mantel tests correlating genetic and geographic distances for each clade separately. A more elegant solution, however, would be to analyze this correlation within clades or groups, so that both pairwise genetic and geographic distances would be standardized within groups before the analyses. The single matrix correlation coefficient obtained in this way would reflect an average effect combined across the different clades, although different sample sizes and local effects should be taken into account. But this will assume that the two processes occur as nested hierarchical effects, without interactions. Another possibility is to use Mantel tests under a multiple correlation and regression design (Manly, 1986; Legendre and Legendre, 1998; Hawkins et al., 2003) to simultaneously evaluate the effect of long-term historical divergence and the effect of more recent and local IBD. In this case, it would be possible to establish which part of the total explained variance of genetic distances could be attributed to these effects and to the overlap between them. These relative values could be obtained simply by performing Mantel tests, using each effect separately and combined into a single model, as described below. Using the notation by Legendre and Legendre (1998), the unexplained variation in genetic divergence (d) is given by 1 - RT2, where RT2 is the squared correlation coefficient of a Mantel test performed using a general linear model that includes both matrices (geographic distances, to evaluate IBD, and the binary model matrix representing long-term historical divergence), which corresponds to the portion (a + b + c). The overlap between IBD and long-term historical divergence (b) is equal to (a + b) + (b + c) - (a + b + c), where (a + b) is given by the R2 of the Mantel test using geographic distances only (R2I), and (b + c) is given by the R2 of the Mantel test using model matrix (R2H). We can then partition variation explained by IBD only (a) and the long-term historical divergence only (c), simply by (R2I - b) and (R2H - b), respectively. APPLICATION: GENETIC DIVERGENCE AMONG LOCAL POPULATIONS OF EUGENIA DYSENTERICA IN CENTRAL BRAZIL We applied these analyses to explain spatial patterns in genetic distances among 10 local populations of Eugenia dysenterica in Goiás State, Central Brazil (Telles et al., 2001a,b, 2003; Diniz-Filho and Telles, 2002). The material analyzed consisted of 112 families obtained from sampling in 10 localities (Figure 1). Leaves of 704 individuals, descendents from these progenies, were submitted to starch gel electrophoresis to evaluate enzymatic patterns. Variation was evaluated at eight polymorphic loci from six isozyme systems, including malate dehydrogenase, alpha esterase (2 loci), 6-phosphogluconate dehydrogenase, phosphoglucomutase (2 loci), phosphoglucoisomerase, and shikimate dehydrogenase. The genetic distances were obtained by Nei’s (1972) coefficients based on eight loci with 34 alleles.
A detailed analysis of the genetic diversity within and among local populations of E. dysenterica was recently conducted by Telles et al. (2003), combining techniques of multivariate and spatial data analyses (both spatial autocorrelation and methods to search for genetic discontinuities, such as the Monmonier algorithm and “wombling” - see Manel et al., 2003). The large genetic divergence between the eastern and western groups of populations, separated by a drop of altitude in the region of Corumbá River valley, cannot be unequivocally associated with a genetic discontinuity (a boundary), in such a way that the most parsimonious explanation for divergence is IBD. However, there are slight differences in the correlogram profiles when the eastern group is analyzed separately, suggesting that different processes could be involved in genetic divergence at smaller and larger geographic distances (Telles et al., 2003) (see Figure 1). Thus, this dataset may be useful to show the application of a partial Mantel test to partition contemporary and historical effects on genetic divergence among populations. The simple matrix correlation between genetic and geographic distance was 0.717 (P < 0.001 under 5,000 permutations); consequently, about 52% of the variation in genetic distances can be attributed to geographic distances between pairs of populations. Repeating the Mantel test with only the other eight populations on the eastern side of the river valley barrier decreased the coefficient from 0.717 to 0.246 (P = 0.12 with 5,000 permutations); so, the relationship between these populations and those on the western side of the barrier is not fully additive and these groups are probably subject to different evolutionary processes. Using a smaller distance class in the eastern group, the matrix correlation was 0.548 (P = 0.007); so, the closest populations within this group (situated less than 56 km from each other - see Telles et al., 2003) are still quite similar, even though the overall relationship between genetic and geographic distance may be low, as expected under IBD. A within-group estimate (standardizing genetic distances within groups) was not indicated in this example because the western group had only two local populations. The binary model matrix designed to separate eastern and western groups was also highly significantly correlated with Nei’s genetic distances, as expected, since in fact the two populations that were spatially isolated from the others by the presumed barrier were also very different based on isozymes (r = 0.848; P < 0.001 with 5,000 permutations). So, long-term divergence explains 72% of the variation in genetic distances (though the information about “history” is quite crude). However, since the long-term divergence binary matrix is also structured in geographic space (r = 0.752; P < 0.001), we cannot interpret these two results independently. We must combine the two matrices (geographic distances and the binary model matrix) as predictors of the genetic distances using a Mantel test, based on a multiple regression design. The R2T of this full model was equal to 73.3%, and using the equations described above, it was possible to determine that, in this case, 21.9% of the variation in the genetic distances can be attributed to the long-term historical divergence alone, while only 1.5% of the variation in genetic distances can be attributed to relationships within groups, due to geographically structured gene flow counteracting genetic drift, as in a contemporary IBD model. In other words, spatial patterns in genetic distances are much better explained by differences between eastern and western groups of populations than by similarity among adjacent local populations within these groups. However, as expected, there is a large overlap between the two processes, in such a way that it is not possible to entirely partition population divergence between historical and contemporary processes (Figure 2).
So, in this data set, genetic divergence is not well explained by IBD alone, as suggested by a simple Mantel test. Although it is not possible to interpret the process independently, and better geographical data are necessary to test the effectiveness of the east-west historical barrier, a long-term historical (and also geographical) divergence may be a more powerful explanation for genetic divergence. Of course, the historical barriers would be more complex, with alternative scenarios at local scales separating other groups of populations in the eastern group. Indeed, phylogenetic correlograms suggest strong autocorrelation effects at short geographic distances (see Telles et al., 2001a, 2003; Epperson, 2003). Also, it is difficult to define the historical scale of IBD processes in relation to the time-scale of a specific barrier, so that the overlap between their explanatory abilities should indeed be relatively high, as we found here. The important point, in this case, it to define the relative magnitude of the two processes, and partial and multiple Mantel test can provide a useful tool in these cases. ACKNOWLEDGMENTS We thank Alexandre S.G. Coelho, Lázaro José Chaves, R. Vencovsky, and Fabrízio D’Ayala Valva for discussions on the geographic population structure of Eugenia dysenterica. The database used for the analyses discussed here was obtained under support of the PRIPE program (Programa Integrado de Pesquisa: Domesticação de Plantas do Cerrado) of CNPq and FINEP/UFG. M.P.C. Telles was supported by a masters fellowship from the Fundação de Apoio à Pesquisa da Universidade Federal de Goiás (FUNAPE/UFG) and by grants from the Vice-Reitoria de Pós-Graduação (VPG/UCG). J.A.F. Diniz-Filho received a CNPq 1B productivity fellowship. REFERENCES Avise JC (2000). Phylogeography. Harvard University Press, Cambridge, MA, USA. Castellano S and Balletto E (2002). Is the partial Mantel test inadequate? Evol. Int. J. Org. Evol. 56: 1871-1873. Congdon BC, Piatt JF, Martin K and Friesen VL (2000). Mechanisms of population differentiation in marbled murrelets: historical versus contemporary processes. Evol. Int. J. Org. Evol. 54: 974-986. Diniz-Filho JAF (2000). Métodos filogenéticos comparativos. Holos, Ribeirão Preto, SP, Brazil. Diniz-Filho JAF and Telles MPC (2002). Spatial autocorrelation analysis and the identification of operational units for conservation in continuous populations. Conserv. Biol. 16: 924-935. Epperson BK (2003). Geographical genetics. Princeton University Press, Princeton, NJ, USA. Hawkins BA, Porter EE and Diniz-Filho JAF (2003). Productivity and history as predictors of the latitudinal diversity gradient of terrestrial birds. Ecology 84: 1608-1632. Lampert KP, Rand AS, Mueller UG and Ryan MJ (2003). Fine-scale genetic pattern and evidence for sex-biased dispersal in the tungara frog, Physalaemus pustulosus. Mol. Ecol. 12: 3325-3334. Legendre P and Legendre L (1998). Numerical ecology. Elsevier, Amsterdam, Holland. Maes GE and Volckaert FAM (2002). Clinal genetic variation and isolation-by-distance in the European eel Anguilla anguilla L. Biol. J. Linn. Soc. 77: 509-521. Manel S, Schwartz MK, Luikart G and Taberlet P (2003). Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 15: 290-295. Manly BFJ (1985). The statistics of natural selection. Chapman & Hall, London, UK. Manly BFJ (1986). Randomization and regression methods for testing for associations with geographical, environmental and biological distances between populations. Res. Popul. Ecol. 28: 201-218. Manly BFJ (1997). Randomization, bootstrap and Monte Carlo methods in biology. Chapman & Hall, London, UK. Nei M (1972). Genetic distance between populations. Am. Nat. 106: 283-292. Raufaste N and Rousset F (2001). Are partial Mantel tests adequate? Evol. Int. J. Org. Evol. 55: 1703-1705. Rodrigues FM, Diniz-Filho JAF, Bataus LAM and Bastos RP (2002). Hypothesis testing of genetic similarity based on RAPD data using Mantel test and model matrices. Genet. Mol. Biol. 25: 435-439. Rousset F (2002). Partial Mantel tests: reply to Castellano and Balletto. Evolution 56: 1874-1875. Santos S, Schneider H and Sampaio I (2003). Genetic differentiation of Macrodon ancylon (Sciaenidae, Perciformes) populations in Atlantic coastal waters of South America as revealed by mtDNA analysis. Genet. Mol. Biol. 26: 151-161. Shipley B (2002). Cause and correlation in biology. Cambridge University Press, Cambridge, UK. Smouse PE, Long JC and Sokal RR (1986). Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 35: 627-632. Sokal RR (1979). Testing statistical significance of geographic variation patterns. Syst. Zool. 28: 227-232. Sokal RR and Rohlf FJ (1995). Biometry. 3rd edn. W.H. Freeman, New York, NY, USA. Sokal RR, Oden NL, Walker J and Waddle DM (1997). Using distance matrices to choose between competing theories and an application to the origin of modern humans. J. Hum. Evol. 32: 501-522. Swofford DL, Olsen GJ, Waddell PJ and Hillis DM (1996). Molecular systematics. In: Phylogeny inference (Hillis DM, Moritz C and Marble BK, eds.). Sinauer Associates, Sunderland, MA, USA, pp. 407-514. Telles MPC and Diniz-Filho JAF (2000). Estimativa da divergência populacional pelas estatísticas F: efeito da autocorrelação nas freqüências alélicas e o teste de Lewontin-Krakauer. Rev. Cienc. Biol. Meio Ambiente PUC/SP 2: 175-188. Telles MPC, Diniz-Filho JAF, Coelho ASG and Chaves LJ (2001a). Autocorrelação espacial das freqüências alélicas em subpopulações de Cagaiteira (Eugenia dysenterica DC, Myrtacea) no sudeste de Goiás. Rev. Bras. Bot. 24: 145-154. Telles MPC, Silva RSM, Chaves LJ, Coelho ASG et al. (2001b). Divergência entre subpopulações de cagaiteira (Eugenia dysenterica) em resposta a padrões edáficos e distribuição espacial. Pesqui. Agropecu. Bras. 36: 1387-1394. Telles MPC, Coelho ASG, Chaves LJ, Diniz-Filho JAF et al. (2003). Genetic diversity and population structure of Eugenia dysenterica DC (“Cagaiteira” - Myrtaceae) in Central Brazil: spatial analysis and implications for conservation and management. Conserv. Genet. 4: 685-695. |
|