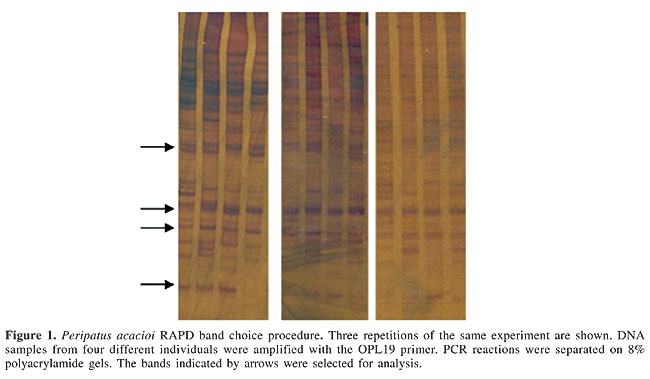
ABSTRACT. RAPD (random amplification of polymorphic DNA) molecular markers can be utilized for analyzing genetic variability in populations for which only a few or no molecular markers are available. They were used in a study of an endangered species, Peripatus acacioi, found in the Tripuí Ecological Station, in Ouro Preto, MG, Brazil. The ecological station was specifically created to protect this velvet worm species, the first of this group found in Brazil. For an initial evaluation of the genetic diversity of this species, DNA samples from the lobopods of four individuals, collected at random, were analyzed using RAPD. Each reaction was run with a different primer (Operon RAPD 10-mer Kits), totaling 13 primers (OPC2, OPC3, OPC4, OPC6, OPC8, OPC10, OPC11, OPL2, OPL7, OPL11, OPL13, OPL18, and OPL19). Due to the low amplification yield, RAPD fragments were separated in polyacrylamide gels and stained with silver nitrate. Numerous bands were observed. Fifty-five of the amplified bands proved to be reproducible, both in terms of presence and intensity. Among these, 27 were variable and 28 were constant. The average number of bands per gel was 4.2. Nine of the 13 primers tested allowed the identification of constant and variable bands among these four individuals. RAPD analysis of genetic variation using silver-stained polyacrylamide gel electrophoresis provided measures of band sharing among the individuals, and therefore could be used in population genetics studies of P. acacioi. Key words: Onychophora, Peripatus acacioi, RAPD INTRODUCTION The phylum Onychophora comprises animal species that have peculiar evolutive characteristics. Aysheaia fossils from the Cambrian period are morphologically similar to contemporary species (Min et al., 1998; Monge-Najera and Hou, 2000). Due to characteristics in common with annelids and arthropods this phylum has been considered intermediate between these two groups. These animals, commonly known as velvet worms, may be ancestral to the arthropods. Recent studies suggested that the Onychophora phylum probably diverged early in arthropod radiation (Herbert et al., 1991). The Onychophora comprise seven genera and approximately a hundred species. They are divided into two families: Peripatidae, found in equatorial regions: Northeastern and Southeastern South America, Eastern Africa, Southeast Asia, and Central America and Peripatopsidae in subtropical regions, such as Chile, Southern Africa, Australia, and New Zealand. The phylum is discontinuous, but the same genus can be found in different continents, such as Opisthopatus, found in Chile and South Africa, and Mesoperipatus, encountered in the Congo and in Central America. This evidence provides additional support for the continental drift theory (Hickman, 1973; Barnes, 1984). Peripatus acacioi, found near the city of Ouro Preto, MG, was the first Onychophora species described in Brazil (Marcus and Marcus, 1955). In 1978, in order to protect this species, an ecological reservation was established, the “Estação Ecológica do Tripuí”. Peripatus acacioi is considered an endangered species (Wieloch, 1998). In addition to Peripatus acacioi, some other Onychophora species have been described in Brazil, which were found in the States of Goiás (Peripatus evelinae), Espírito Santo (Peripatus sp), Pará (Peripatus edwardsii and Peripatus tucupi), and Mato Grosso (Peripatus heloisae) (Carvalho, 1941; Froehlich, 1968; Wieloch, 2001). Peripatus acacioi has been the subject of several studies, including a brief description of its karyotype and DNA content (Schreiber and Cavenaghi, 1971). Morphological and physiological aspects of this species have also been studied (Campiglia, 1976; Hoyle and Del Castillo, 1979; Bicudo and Campiglia, 1985; Lane and Campiglia, 1987; Silva et al., 2000). Genetic structure and phylogenetic analyses of Onychophora populations in Australia and New Zealand were made with isozymes, mitochondrial DNA, and microsatellites (Gleeson et al., 1998; Sunnucks and Wilson, 1999; Trewick, 1998, 1999, 2000; Rockman et al., 2001). High levels of genetic diversity were reported in most of these studies. In addition, the existence of new species has been suggested. Based on behavioral characteristics of the species, such as a preference for humid and dark environments and limited locomotion of the individuals, P. acacioi populations in the Tripuí Ecological Station were expected to be structured in small and relatively isolated subpopulations. The use of RAPD (random amplification of polymorphic DNA) was chosen to test this hypothesis. RAPD amplification occurs if the primer anneals to DNA at intervals shorter than 4,000 bp and in opposite orientations. Each oligonucleotide can recognize several sequences in the genome, resulting in the amplification of many fragments (Williams et al., 1990; Welsh and McClelland, 1990). Beyond being simple, fast and low in cost, RAPD does not require previous knowledge of DNA sequences and can be done with small amounts of DNA. Another advantage is that a set of arbitrary primers can be utilized for any organism. However, RAPD has also some disadvantages, including a high susceptibility to PCR conditions, which results in low reproducibility (Hadrys et al., 1992; Liu and Cordes, 2004). RAPD can be used in the analysis of gene flow, hybrid speciation, paternity investigation, and kin relationships, and even in sample admixtures (Hadrys et al., 1992). Usually, fragments amplified by RAPD are visualized in ethidium bromide-stained agarose gels, although the use of polyacrylamide gels stained with silver nitrate has occasionally been described (Vidigal et al., 1994; Abdel-Hamid et al., 1999; Barnini et al., 2004). Due to the extremely low PCR yield of P. acacioi DNA, electrophoretic separation in polyacrylamide gels stained with silver nitrate was chosen in order to evaluate the applicability of RAPD for this species. We developed this study to examine the genetic diversity of P. acacioi in the “Estação Ecológica do Tripuí”. MATERIAL AND METHODS DNA samples from four randomly chosen individuals (P1-4) were used to test the efficiency of the use of RAPD molecular markers in P. acacioi specimens. The animals were collected from February to April, 2000, using permission 001/00 from “Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis”. Due to legal restrictions for collecting material from endangered species, only two lobopods from each individual were collected. The animals were anesthetized with CO2 and the paws were sampled using a forceps and a scalpel. The collected material was stored in 70% ethanol. The Wizard® Genomic DNA Purification Kit (Promega) was used for DNA extraction. Due to the reduced volume of the final solution (25 µL) the DNA was not quantified. The PCR reactions were run with 1:120 dilutions. RAPD reactions were run with primers from the Operon RAPD 10-mer Kit, KitC: OPC2, OPC3, OPC4, OPC6, OPC8, OPC10, OPC11, and KitL: OPL2, OPL6, OPL7, OPL11, OPL13, OPL18, and OPL19. PCR reactions were done in 20 µL of a solution containing: 10 mM Tris-HCl, pH 8.0, 50 mM KCl, 2 mM MgCl2, 0.1 mM of each dNTP, 0.25 µM primer, 1 U Taq DNA polymerase, and 2 µL genomic DNA (from the 1:120 dilution). All reactions were done at least twice (up to four times) for analysis of banding pattern reproducibility. PCR reactions were run in a PTC-100TM thermocycler (M.J. Research, Inc.), using the following program: 1 min denaturation step at 95°C, followed by 35 cycles of 1 min at 95°C, 10 s at 36°C, and 2 min at 72°C, and a 7 min last extension step at 72°C. RAPD products were electrophoresed on 8% polyacrylamide gels (PAGE), subsequently stained with silver nitrate. Thirteen of the 14 oligonucleotides that were tested yielded products that were well visualized on polyacrylamide gels and were thus selected for analysis (OPC2, OPC3, OPC4, OPC6, OPC8, OPC10, OPC11, OPL2, OPL7, OPL11, OPL13, OPL18, and OPL19). RESULTS The first question addressed was the reproducibility of the separation of RAPD products on silver-stained PAGE. For these analyses, we compared the results obtained for each individual in two to four gels of each RAPD system. Only those bands that were present in all gels with similar intensity, considering the same individuals and the same oligonucleotide, were scored (see Figure 1).
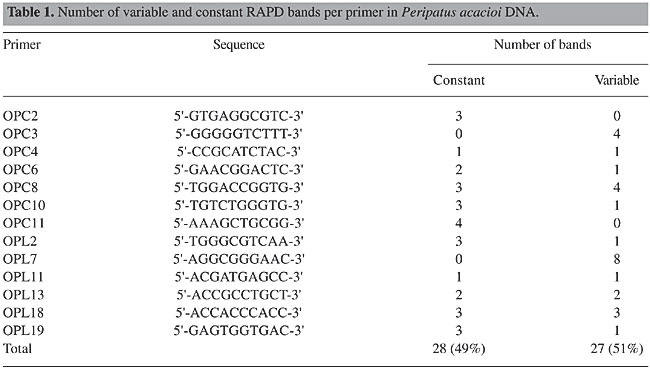
In addition to reproducibility, the total number of bands, number of constant bands (bands present in the four individuals), and the number of variable bands (bands present in some individuals and absent in others) were evaluated for each oligonucleotide (Table 1). The total number of reproducible bands per gel ranged from two to eight, averaging 4.2 bands/gel. Some of the oligonucleotides yielded only constant bands (OPC2 and OPC11), while others rendered only variable bands (OPC3 and OPL7).
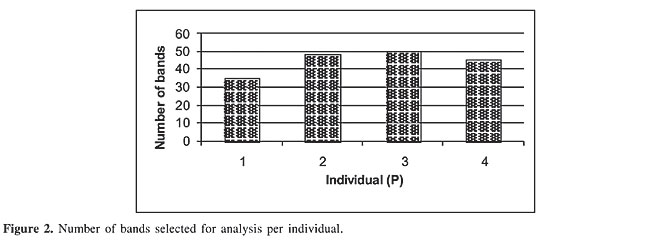
The average number of bands per individual was 44.5, ranging from 35 to 50 (Figure 2).
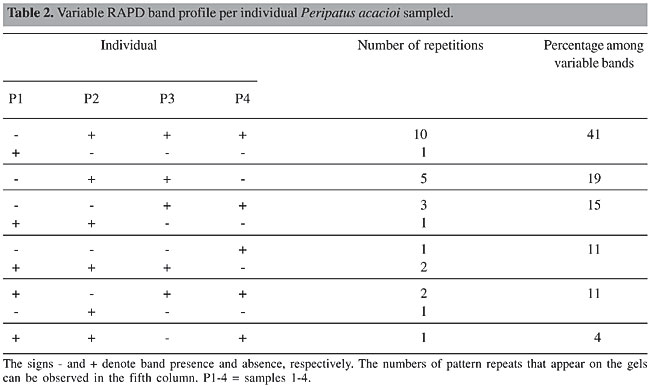
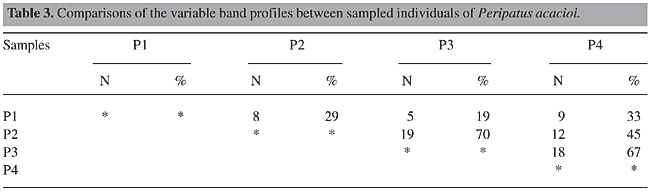
Twenty-seven of the 55 bands selected for the analysis were variable, that is they were not present in all the individuals. Highly similar RAPD band patterns were observed in individuals P2 and 3 (19 shared bands, 70%) (See Table 2). The band patterns were compared between paired individuals (Table 3). Individual P1 was the most different among the individuals.
DISCUSSION RAPD is a valuable tool for the analysis of the genetic diversity of populations, whenever specific genetic markers are not available. RAPD is usually analyzed on agarose gels. However, since the PCR yield of P. acacioi DNA was low, the PCR products were separated and visualized on silver nitrate-stained PAGE. This facilitated visualization of the amplified material and identification of the reproducible bands; however, an excessive number of bands was observed on the gel, because of the high sensitivity of the method. Silver-stained PAGE has seldom been used for separation of RAPD fragments. Stift et al. (2003), in a study of Cucurbita species, compared RAPD fragment separation on agarose gels and PAGE. They concluded that the separation of RAPD products on PAGE instead of agarose has some advantages, including a larger number of visible bands, cleaner band separation, lower DNA and PCR reagent requirements, better cost efficiency, due to the lower PAGE price when compared to agarose, avoidance of handling of ethidium bromide, which is a mutagenic substance, and the possibility of indefinite storage of the original PAGE. Barnini et al. (2004), in a study of Pseudomonas aeruginosa, separated the same RAPD-PCR reactions with ethidium bromide-stained agarose gels and with silver-stained PAGE. The latter method gave rise to a much larger number of bands. This allowed a finer discrimination among bacterial amplification profiles. Besides the large number of RAPD bands generated with silver-stained PAGE, many of the bands are not consistently reproducible, i.e., they are not present in all repetitions of the same experiment. However, in our study, it was possible to identify bands that were always present, and with similar intensity, when the same individual and the same set of primers were considered. The criteria were fulfilled by 55 bands, with an average of 4.2 valid bands per gel (variation interval from 2 to 8 valid bands per gel). Approximately half of the bands that were scored were variable (27 of 55 bands). The ability to analyze interindividual variation is an important criterion for the selection of a genetic marker. By this criterion, the best primer systems were OPL7, OPC3, OPC8, and OPC18, which gave 8, 4, 4, and 3 variable bands each, respectively. Variation between the tested individuals was not observed when the OPC2 and OPC11 markers were used. Although these markers may not add further information on genetic variability in this sample, they may still be useful, for example, if they lead to the identification of a species-specific band pattern. However, due to the small number of sampled individuals, conclusions about these primers cannot be made. RAPD has been used in the evaluation of genetic diversity in several organisms. The systems utilized have varied considerably from one study to another. In genetic diversity studies on two Oleria onega subspecies of butterflies, seven oligonucleotides were chosen, which allowed the amplification of 88 variable bands (Gallusser et al., 2004). In a Hemipteran, Marchalina hellenica, 10 oligonucleotides amplified 35 bands (Margaritopoulos et al., 2003), and in studies on the locust species, Rhammatocerus schistocercoides, 11 oligonucleotides yielded 90 bands, of which 63 were variable (Silva et al., 2002). Considering the small number of individuals in our study and the relatively large number of variable bands observed, we believe that RAPDs will allow the evaluation of genetic diversity in P. acacioi. Another use for the RAPD technique is the determination of taxonomic identity based on the observation of constant bands shared only by individuals of the same genus or species. These bands are considered genus- or species-specific markers (Hadrys et al., 1992). RAPD could be useful for distinguishing animal species with conserved morphology, such as those of the phylum Onychophora. RAPD systems have been used in the characterization of species and subspecies of the insects Cochliomyia hominivorax and C. macellaria (Skodal et al., 2002), the prune species Prunus salicina, Prunus domestica and Prunus cerasefera (Bianchi et al., 2003), peaches and nectarines of the species Prunus persica (Lima et al., 2003), pears of the species Pyrus ssp (Sawazaki et al., 2002), rattle snakes of the subspecies Crotalus durissus collilineatus and Crotalus durissus terrificus (Echeverrigaray et al., 2001), and cattle of the breed Crioulo Lageano (Spritze et al., 2003). However, in some cases genetic marker systems based on RAPDs only allow individual identification. For example, individual variation was observed in the Triatominae species, Triatoma longipennis, Triatoma picturata, and Triatoma pallidipennis, but species-specific bands were not detected (Brenière et al., 2003). This is the first time that the applicability of RAPD has been tested to solve questions of population genetic structure of Peripatus. Studies based on mitochondrial DNA of New Zealand Onychophora populations pointed to the existence of a great number of cryptic species (Trewick, 1998, 1999, 2000). CONCLUSIONS We propose some criteria for the analysis of genetic variation based on RAPD separation in silver-stained PAGE: 1. Due to the high variation observed among different PCR reactions, conclusions should be based on at least two independent PCR reactions per individual. 2. Only those bands produced by the same oligonucleotide primer that show similar intensities in all repetitions of the same individual should be scored. 3. Electrophoretic conditions should be kept constant among different runs. Although only four individuals were tested, some conclusions relevant for future investigations on the genetic variability of Peripatus acacioi could be made. Oligonucleotide primers OPC2, OPC3, OPC4, OPC6, OPC8, OPC10, OPC11, OPL2, OPL7, OPL11, OPL13, OPL18, and OPL19 allowed the identification of both constant and variable reproducible bands. Oligonucleotide primers OPC2 and OPC11 rendered only constant bands (i.e., they were present in all four individuals). Only variable bands were observed with primers OPC3 and OPL7. Twenty-seven of a total of 55 reproducible bands were variable. The methodology that was employed demonstrated band sharing among individuals, thus being applicable to studies on the genetic structure of populations of this species. ACKNOWLEDGMENTS Research supported by the Brazilian National Research Council (CNPq) grant No. 46.9539/00-3. D.M. DeLaat was supported by a fellowship from Coordenadoria de Aperfeiçoamento de Pessoal de Ensino Superior (CAPES). Authorization for sampling Peripatus acacioi individuals was granted by the Instituto Brasileiro do Meio Ambiente (IBAMA), Instituto Estadual de Florestas, Estação Ecológica do Tripuí, and Mr. Aristides S.G. Neto. Special thanks go to Prof. Alfredo Hannemann Wieloch (UFMG) and Dr. Sérvio Pontes Ribeiro (UFOP) for helping with P. acacioi sampling. We also thank Ms. Joana Bernardes de Assis for reviewing this manuscript. REFERENCES Abdel-Hamid Z, Molfetta JB, Fernandez V and Rodrigues V (1999). Genetic variation between susceptible and non-susceptible snails to Schistosoma infection using random amplified polymorphic DNA analysis (RAPDs). Rev. Inst. Med. Trop. São Paulo 41: 291-295. Barnes RD (1984). Zoologia dos invertebrados. 4th edn. Roca, São Paulo, SP, Brazil. Barnini S, Dodi C and Campa M (2004). Enhanced resolution of random amplified polymorphic DNA genotyping of Pseudomonas aeruginosa. Lett. Appl. Microbiol. 39: 274-277. Bianchi VJ, Fachinello JC and Schuch MW (2003). RAPDs na caracterização genético-molecular e no estudo da variabilidade genética de cultivares de ameixeira. Rev. Bras. Frutic. 25: 272-274. Bicudo JE and Campiglia S (1985). A morphometric study of the tracheal system of Peripatus acacioi Marcus and Marcus (Onychophora). Respir. Physiol. 60: 75-82. Brenière SF, Taveira B, Bosseno MF, Ordoñez R et al. (2003). Preliminary results of random amplification of polymorphic DNA among Triatominae of the phyllosoma complex (Hemiptera, Reduviidae). Mem. Inst. Oswaldo Cruz 98: 1033-1038. Campiglia SS (1976). The blood of Peripatus acacioi Marcus and Marcus (Onychophora). III. The ionic composition of the hemolymph. Comp. Biochem. Physiol. A. 54: 129-133. Carvalho AL (1941). Sobre Peripatus heloisae do Brasil central. Bol. Mus. Nac. Rio J. Zool. 2: 57-73. Echeverrigaray S, Grazziotin G and Grazziotin F (2001). Random amplified polymorphisms between two South American subspecies of rattlesnakes (Crotalus durissus collilineatus and Crotalus durissus terrificus). Braz. Arch. Biol. Technol. 44: 313-317. Froehlich CG (1968). On some Brazilian onychophores. Beitr. Neotrop. Fauna 5: 160-171. Gallusser S, Guadagnuolo R and Rahier M (2004). Genetic (RAPD) diversity between Oleria onega agarista and Oleria onega ssp. (Ithomiinae, Nymphalidae, Lepidoptera) in northeastern Peru. Genetica 121: 65-74. Gleeson DM, Rowell DM, Tait NN, Briscoe DA et al. (1998). Phylogenetic relationships among onychophora from Australasia inferred from the mitochondrial cytochrome oxidase subunit I gene. Mol. Phylogenet. Evol. 10: 237-248. Hadrys H, Balick M and Schierwater B (1992). Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol. Ecol. 1: 55-63. Hebert PDN, Billington N, Finston TR, Boileau MG et al. (1991). Genetic variation in the onychophoran Plicatoperipatus jamaicensis. Heredity 67: 221-229. Hickman CP (1973). Biology of the invertebrates. 2nd edn. Mostry, Saint Louis, MO, USA. Hoyle G and Del Castillo J (1979). Neuromuscular transmission in Peripatus. J. Exp. Biol. 83: 13-29. Lane NJ and Campiglia SS (1987). The lack of a structured blood-brain barrier in the onychophoran Peripatus acacioi. J. Neurocytol. 16: 93-104. Lima MR, Augustin E and Choer E (2003). Caracterização de cultivares de pessegueiro e de nectarineira por marcadores moleculares. Pesqui. Agropecu. Bras. 38: 349-355. Liu ZJ and Cordes JF (2004). DNA marker technologies and their applications in aquaculture genetics. Aquaculture 238: 1-37. Marcus E and Marcus E (1955). A new peripatus from Minas Gerais, Brazil. An. Acad. Bras. Cienc. 27: 189-193. Margaritopoulos JT, Bacandritsos N, Pekas AN, Stamatis C et al. (2003). Genetic variation of Marchalina hellenica (Hemiptera: Margarodidae) sampled from different hosts and localities in Greece. Bull. Entomol. Res. 93: 447-453. Min GS, Kim SH and Kim W (1998). Molecular phylogeny of arthropods and their relatives: polyphyletic origin of arthropodization. Mol. Cells 8: 75-83. Monge-Najera J and Hou X (2000). Disparity, decimation and the Cambrian “explosion”: comparison of early Cambrian and present faunal communities with emphasis on velvet worms (Onychophora). Rev. Biol. Trop. 48: 333-351. Rockman MV, Rowell DM and Tait NN (2001). Phylogenetics of Planipapillus, lawn-headed onychophorans of the Australian Alps, based on nuclear and mitochondrial gene sequences. Mol. Phylogenet. Evol. 21: 103-116. Sawazaki HE, Barbosa W and Colombo CA (2002). Caracterização e identificação de cultivares e seleções de pereiras através de marcadores RAPD. Rev. Bras. Frutic. 24: 447-452. Schreiber G and Cavenaghi TMCM (1971). Variations of DNA content during the phylogenesis. Riv. Istochim. Norm. Patol. Pavia 17: 53-59. Silva JBT, Tigano MS, Magalhães BP and Cordeiro CMT (2002). Polymorphism of the grasshopper Rhammatocerus schistocercoides populations revealed by RAPD. Pesqui. Agropecu. Bras. 37: 1669-1673. Silva JR, Coelho MP and Nogueira MI (2000). Induced inflammatory process in Peripatus acacioi Marcus et Marcus (Onychophora). J. Invertebr. Pathol. 75: 41-46. Skodal SR, Pornkulwat S and Foster JE (2002). Random amplified polymorphic DNA markers for discriminating Cochliomyia hominivorax from C. macellaria (Diptera: Calliphoridae). Bull. Entomol. Res. 92: 89-96. Spritze A, Egito AA and Mariante AS (2003). Caracterização genética da raça bovina Crioulo Lageano por marcadores moleculares RAPD. Pesqui. Agropecu. Bras. 38: 1157-1164. Stift G, Pachner M and Lelley T (2003). Comparison of RAPD fragment separation in agarose and polyacrylamide gel by studying Cucurbita species. Cucurbit Genet. Coop. Rep. 26: 62-65. Sunnucks P and Wilson AC (1999). Microsatellite markers for the onychophoran Euperipatoides rowelli. Mol. Ecol. 8: 899-900. Trewick SA (1998). Sympatric cryptic species in New Zealand Onychophora. Biol. J. Linn. Soc. 63: 307-329. Trewick SA (1999). Molecular diversity of Dunedin peripatus (Onychophora: Peripatopsidae). N. Z. J. Zool. 26: 381-393. Trewick SA (2000). Mitochondrial DNA sequences support allozyme evidence for cryptic radiation of New Zealand Peripatoides (Onychophora). Mol. Ecol. 9: 269-282. Vidigal THDA, Neto ED, Carvalho OS and Simpson AJG (1994). Biomphalaria glabrata: extensive genetic variation in Brazilian isolates revealed by random amplified polymorphic DNA analysis. Exp. Parasitol. 79: 187-194. Welsh J and McClelland M (1990). Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 18: 7213-7218. Wieloch AH (1998). Peripatus acacioi Marcus and Marcus, 1995. In: Livro vermelho das espécies ameaçadas de extinção da fauna de Minas Gerais (Machado ABM, Fonseca GAB, Machado RB, Aguiar LMS et al., eds.). Fundação Biodiversitas, Belo Horizonte, MG, Brazil, pp. 567-568. Wieloch AH (2001). Biologia populacional de Peripatus acacioi Marcus and Marcus, 1955 (Onychophora) da Estação Ecológica do Tripuí, MG. Relatório final. Fapemig, Belo Horizonte, MG, Brazil. Williams JGK, Kubelik AR, Livak KJ, Rafalski JA et al. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18: 6531-6535. |
|