ABSTRACT. Various chromosomal banding techniques were utilized on the catfish, Iheringichthys labrosus, taken from the Capivara Reservoir. C-banding regions were evidenced in telomeric regions of most of the chromosomes. The B microchromosome appeared totally heterochromatic. The restriction endonuclease AluI produced a banding pattern similar to C-banding in some chromosomes; the B microchromosome, when present, was not digested by this enzyme and remained stained. G-banding was conspicuous in almost all the chromosomes, with the centromeres showing negative G-banding. When the restriction endonuclease BamHI was used, most of the telomeres remained intact, while some centromeres were weakly digested. The B chromosome was also not digested by this enzyme. The first pair of chromosomes showed a pattern of longitudinal bands, both with G-banding and BamHI; this was more evident with G-banding. This banding pattern can be considered a chromosomal marker for this population of I. labrosus. Key words: Iheringichthys labrosus, Restriction endonucleases, C-banding, G-banding INTRODUCTION As the considerable chromosomal diversity in fish becomes better known, it has become clear that various methods, both basic and more advanced, are necessary for an adequate cytogenetic characterization of Neotropical fish. Diploid number, chromosome formula and chromosomal banding, from the simplest techniques to those that provide high resolution and specificity, have become very important as cytogenetic markers for understanding chromosome diversity in Neotropical fish. These markers facilitate the pairing of homologs, highlight differences between apparently similar karyotypes, and can even reveal mechanisms of chromosome rearrangements (Ozouf-Costaz and Foresti, 1992; Almeida-Toledo, 1998). The use of C-banding techniques in fish has made it possible to detect heterochromatin regions, to correctly identify homologous chromosomes, and even to identify supernumerary or B chromosomes. G-banding reveals euchromatic bands in chromosomes, although the exact mechanisms by which G bands are produced have still not been completely elucidated. An alternative chromosomal banding technique involves the use of restriction endonucleases. These restriction endonucleases can produce banding patterns that are highly specific for each type of enzyme. According to Swarça (2003), the analysis of these patterns facilitates chromosome classification, the differentiation of heterochromatin and the study of chromosomal polymorphisms. We examined the karyotype of Iheringichthys labrosus, a catfish of some commercial importance in Brazil, by means of C- , G-banding and with the restriction endonucleases, AluI and BamHI. MATERIAL AND METHODS Twenty-five specimens of I. labrosus collected in two different municipalities near the Capivara Reservoir, Paraná, Brazil, were collected. There were 15 individuals (9 females and 6 males) from Sertanópolis (PR, Brazil) and 10 individuals (5 females, 4 males and one of undetermined sex) from Porecatu (PR, Brazil). Mitotic chromosome preparations were obtained according to Bertollo et al. (1978). C- and G-banding were performed according to Sumner (1972) and Cano et al. (1996), respectively. Restriction endonucleases were employed according to Sánchez et al. (1990), with some modifications, i.e., AluI (0.3 units/µL, at 37°C for 4 h), BamHI (0.4 units/µL, at 37°C for 15 h). In view of the difficulty in determining chromosome morphology after chromosomal banding, the karyotypes were organized based on size, in decreasing order. RESULTS AND DISCUSSION A previous cytogenetic study of I. labrosus from the Capivara Reservoir (Carvalho and Dias, 2005) showed a diploid number of 56 chromosomes, also indicating the presence of a supernumerary microchromosome, with both inter- and intra-individual variation. C-banding showed heterochromatin in the telomeric regions in most of the chromosomes of I. labrosus from the Capivara Reservoir at the two localities; few chromosomes showed C-banding positive centromeres. When present, the supernumerary or B microchromosome appeared totally heterochromatic (Figure 1A).
Studies of the heterochromatin distribution pattern in other populations of I. labrosus have shown that each population has a characteristic pattern; however, some of them are similar to the evidenced one in the present work. Vissotto et al. (1999), for example, studied a population of I. labrosus from the Jurumirim Reservoir (SP) and found a large quantity of heterochromatin in interstitial and terminal regions. Dias and Foresti (1990) found centromeric and telomeric heterochromatin in a population from the Mogi-Guaçu River (SP), and practically the same result was reported by Garcia et al. (1990) in I. labrosus from the Paraná River. Carvalho et al. (2004) observed that I. labrosus from the Tibagi River (PR) has heterochromatin distributed mainly in telomeric regions, including a strongly heterochromatic pair of telomeres, which was suggested to be a marker for this population. AluI, which identifies and cleaves the DNA specific sequence AG/CT, produced a similar C-banding pattern in some chromosomes. Some telomeres remained intact after enzyme treatment, while others were cleaved, therefore possessing cleavage sites for this enzyme (Figure 1B). Since telomeres are heterochromatic, this suggests that different types of heterochromatin occur in this population. When present, the B microchromosome was not digested by AluI, and it remained stained (Figure 1B). Carvalho et al. (2004) utilized this enzyme in I. labrosus from the Tibagi River, in which it was also possible to see a banding pattern similar to C-banding, in various chromosomes of the complement. Swarça et al. (1999, 2001) also obtained banding patterns similar to C-banding with AluI in Pinirampus pirinampu and Pimelodus maculatus, as did Swarça (2003) in Steindachneridion sp and S. scripta.
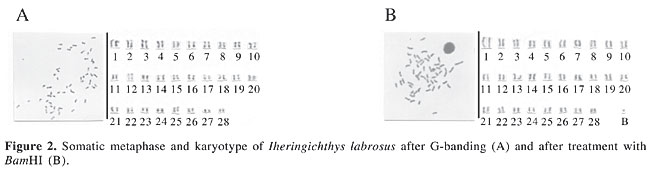
G-banding analysis, with the utilization of trypsin, demonstrated a conspicuous banding pattern in almost all the chromosomes of the complement (Figure 2A). The centromeres for the most part stained weakly. That is, they were negative for G-banding, showing that these regions are sensitive to the proteolytic action of trypsin, while most of the telomeres, which are heterochromatic, displayed strong staining, and were therefore not digested by trypsin. The B microchromosome could not be visualized in the G-banding preparations. In a study of the family Pimelodidae, Swarça (2003) utilized G-banding in chromosome preparations of Steindachneridion sp, S. scripta and Pseudoplatystoma corruscans; she found a pattern of longitudinal chromosomal differentiation in the three species. These findings, taken together with our findings for I. labrosus from the Capivara Reservoir, support the proposition made by Maistro et al. (1999) that the difficulty in detecting G-banding in fish chromosomes is due more to technical problems than to the lack of structural compartmentalization in the genome of this fish. With the utilization of BamHI, which identifies and cleaves the DNA sequence G/GATCC, most of the telomeres of I. labrosus appeared intact, and some centromeres were slightly digested, thereby displaying little staining. The B chromosome was not digested by this enzyme (Figure 2B). In the family Pimelodidae, G-banding patterns have been obtained with this enzyme for Steindachneridion sp and S. scripta (Swarça, 2003). However, in our study, the banding patterns seen with BamHI were not so evident in G-banding. Although the centromeres were well digested by trypsin, they appeared to have few cleavage sites for this restriction enzyme. The first pair of chromosomes possessed a longitudinal banding pattern, with both G-banding and BamHI effects, being more evident in G-banding; thus this characteristic could be considered a chromosomal marker for this population of I. labrosus. The findings obtained here, using different chromosomal banding techniques, provide an integral chromosomal characterization of I. labrosus from the Capivara Reservoir, demonstrating various types of heterochromatin and chromosomes in this species, which can serve as discriminatory cytogenetic markers. These markers permit the differentiation of populations, and they contribute to our knowledge of the cytogenetics of this group of fish. ACKNOWLEDGMENTS The authors are grateful to CAPES for financial support. We are also thankful to Dr. Albert Leyva for his help in the preparation of the manuscript. REFERENCES Almeida-Toledo LF (1998). Cytogenetic markers in Neotropical freshwater fishes. In: Phylogeny and classification of Neotropical fishes (Malabarba LR, ed.). EDIPUCRS, Porto Alegre, RS, Brazil, pp. 583-588. Bertollo LAC, Takahashi C and Moreira-Filho O (1978). Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae). Rev. Bras. Genet. 1: 103-120. Cano J, Pretel A, Menendez S, Garcia S et al. (1996). Determination of early stage of sex chromosome differentiation in the sea bas Dicentrarctus labrax L. (Pisces, Perciformes). Cytobios 87: 45-59. Carvalho RA and Dias AL (2005). Cytogenetic characterization of B chromosomes in two populations of Iheringichthys labrosus (Pisces, Pimelodidae) from the Capivara Reservoir (Paraná, Brazil). Caryologia 58: 269-273. Carvalho RA, Giuliano-Caetano L and Dias AL (2004). Cytogenetic analysis A- and B-chromosomes of Iheringichthys labrosus (Pisces, Pimelodidae) from the Tibagi River, Paraná, Brazil. Cytologia 69: 381-385. Dias AL and Foresti F (1990). Algumas considerações sobre o cariótipo de Iheringichthys labrosus (Siluriformes, Pimelodidae) do rio Mogi-Guaçu. Anais do III Simpósio de Citogenética Evolutiva e Aplicada de Peixes Neotropicais, Botucatu, SP, Brazil, p. 32. Garcia RMG, Sachete R and Martins-Santos IC (1990). Aspectos citogenéticos de Iheringichthys labrosus (Pisces, Pimelodidae) do rio Paraná, região de Porto Rico, PR. Anais do III Simpósio de Citogenética Evolutiva e Aplicada de Peixes Neotropicais, Botucatu, SP, Brazil, p. 33. Maistro EL, Oliveira C and Foresti F (1999). R- and G-band patterns in Astyanax scabripinnis (Pisces, Characiformes, Characidae). Genet. Mol. Biol. 22: 201-204. Ozouf-Costaz C and Foresti F (1992). Fish cytogenetic research: advances, applications and perspectives. Neth. J. Zool. 42: 277-290. Sánchez L, Martínez P, Viñas A and Bouza C (1990). Analysis of the structure and variability of nucleolar organizer regions of Salmo truta by C-, Ag-, and restriction endonuclease banding. Cytogenet. Cell Genet. 54: 6-9. Sumner AT (1972). A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 75: 304-306. Swarça AC (2003). Contribuição à citogenética dos Pimelodidae de grande porte: estudos cariotípicos de 4 espécies do “subgrupo” Sorubiminae. Universidade Federal do Paraná, Curitiba, PR, Brazil. Swarça AC, Giuliano-Caetano L and Dias AL (1999). Cytogenetic characterization through chromosomic banding of Pinirampus pirinampu (Pisces, Pimelodidae) from the Tibagi river basin PR/Brazil. Caryologia 52: 31-35. Swarça AC, Giuliano-Caetano L and Dias AL (2001). Analyses of nucleolus organizer regions and heterochromatin of Pimelodus maculatus (Pisces, Pimelodidae). Genetica 110: 97-100. Vissotto PC, Foresti F and Oliveira C (1999). Supernumerary chromosomes in two species of the family Pimelodidae (Teleostei, Siluriformes). Chromosome Sci. 3: 9-13. |
|