ABSTRACT. Proteases perform a wide variety of functions inside and outside cells, regulating many biological processes. Infectious microorganisms use proteases, either secreted or attached to their cell surface to weaken and invade their hosts. Therefore, proteases are targets for drugs against a diverse set of diseases. Paracoccidioides brasiliensis is the most prevalent fungal pathogen causing systemic mycosis in Latin America. The development of paracoccidioidomycosis depends on interactions between fungal and host components and proteases have been described as important factors implicated in the mechanism of host colonization by fungi. The primary goal for this study is to present an overview of the transcriptome sequences - identified cDNAs that encode proteases. We obtained a total of 53 cDNAs encoding proteases; 15 were classified as ATP-independent, 12 as ATP-dependent, 22 as proteasome subunits, and 4 as deubiquitinating proteases. The mechanisms and biological activity of these proteases differ in substrate specificity and in catalytic mechanisms. Key words: Paracoccidioides brasiliensis, Host-fungus interaction, Proteases, Proteasome INTRODUCTION Proteases are enzymes that cleave proteins catalysing the hydrolysis of peptide bond. Based on their catalytic mechanisms, proteases can be classified into five main classes: i) aspartyl proteases; ii) metalloproteases; iii) cysteine proteases; iv) threonine proteases, and v) serine proteases. Proteases in the first two classes use an activated water molecule as a nucleophile to attack the peptide bond of the substrate, whereas in the last three classes the nucleophile is a catalytic amino-acid residue located in the active site (Rawlings and Barrett, 1993; Barrett et al., 1998). Proteases are also grossly subdivided into two major groups, depending on their site of action. Exopeptidases cleave the peptide bond proximal to the amino or carboxyl termini of the substrate whereas endopeptidases cleave peptide bonds distant from the termini of the substrate (Watson, 1976). Proteolytic enzymes play many physiological roles and are essential factors for homeostatic control in organisms. Proteases are widely produced amongst fungi and serve a number of different roles within fungal systems including nutrient cycling and post-translational processing (North, 1982). In some instances a correlation between protease production and pathogenicity was reported. There is accumulating evidence of the direct involvement of fungi proteases in different phases of the host-fungi interactions. The physiological role of proteases during colonization of the host is thought to be the degradation of the skin and mucosal barriers, digestion of host proteins to provide nutrients and attack the lymphocytes and macrophages, affecting the immune defenses of the host (Hube, 2000; Yang, 2003). A fraction of the proteolytic activity of cells is ATP independent. In addition, studies have identified three ATP-dependent systems involved in protein degradation in eukaryotic organisms (Menon and Goldberg, 1987). These systems include: i) proteases that require the binding and hydrolysis of ATP for proteolytic activity (Lon, ClPs and the 26 S protease have been identified in detail; ii) the ubiquitin conjugating system, and iii) chaperone proteins. Paracoccidioides brasiliensis is a dimorphic fungus that alternates between a mycelium phase in the free environment and a yeast phase in the human host. Primary infection starts in the lungs after inhalation of fungal propagules which then transform into the pathogenic yeast form. Primary infection is usually spontaneously healed; active paracoccidioidomycosis is estimated to develop in approximately 2% of the infected individuals (McEwen et al., 1995). P. brasiliensis thus represents a serious public health challenge, with social and economical importance. The observation that only a percentual of infected individuals can develop the disease points to both the pathogenic potential of P. brasiliensis and the importance of host defense in controlling fungal infection. P. brasiliensis expresses some molecules that account for its ability to evade efficiently the host protective immune system and proteases should be included with these molecules. Protease-like activity in the culture filtrates of P. brasiliensis was originally noted and an exocellular serine protease has been characterized as a molecule that cleaves in vitro the main components of the basal membrane (Carmona et al., 1995; Puccia et al., 1999). Although potentially associated with the invasion process the role of P. brasiliensis proteases in the fungus ability to cause disease remains to be elucidated. The objective of this review is to summarize the information about the transcriptome-based identification of proteases of P. brasiliensis. The availability of primary structural information about a group of the identified expressed sequence tags (ESTs) allows further analysis and a better overall understanding of pathogen interactions with human host. This insight may shed light on the elucidation of specific functions in which these proteins are involved as well as discover the role of these proteins in the pathogenesis of P. brasiliensis. METHODS Annotated ESTs encoding energy-dependent and -independent proteases were obtained in P. brasiliensis Transcriptome Project database (http://www.biomol.unb.br/Pb). We screened available databases of proteases, including MEROPS (http://merops.sanger.ac.uk/) and Pfam (http://pfam.wustl.edu). The search for similarity was conducted using the BLAST search tools, using the interface web of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/). Domains and predicted active sites were screened using the ProfileScan (http://hits.isb-sib.ch/cgi-bin/PFSCAN?) and ScanProsite algorithms (http://ca.expasy.org/tools/scanprosite/). Multiple sequence alignments were generated using the program ClustalX 1.81 software (Thompson et al., 1997). RESULTS AND DISCUSSION Proteases of Paracoccidioides brasiliensis By using the primary information that was retrieved from the P. brasiliensis transcriptome (http://www.biomol.unb.br/Pb), combined with data from the MEROPS database (http://merops.sanger.ac.uk/) we have annotated 53 ORFs encoding energy-independent and -dependent proteases, including proteasome subunits, aspartyl, cysteine, metallo, and serine proteases. These cDNAs that encode protease homologues in the fungus transcriptome were annotated, as shown in Figure 1. The proteases of P. brasiliensis are distributed as following: 5.6% aspartyl proteases, 11.3% cysteine proteases, 22.6% metalloproteases, 18.8% serine proteases, and 41.5% proteasome subunits.
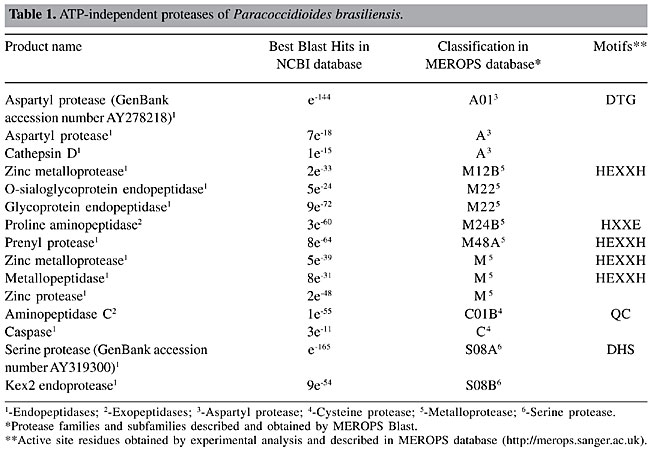
Energy-independent proteases of Paracoccidioides brasiliensis Exopeptidases and endopeptidases We have annotated a total of 15 cDNAs, which encode energy-independent protease homologues in the fungus transcriptome, as shown in Table 1. From these, two were classified as exopeptidases and the remaining 13 as endopeptidases. The energy-independent proteases are distributed as following: three aspartyl, two cysteine, eight metallo, and two serine proteases.
Aspartyl proteases of Paracoccidioides brasiliensis Three aspartyl proteases were found in the P. brasiliensis transcriptome (Table 1, Figure 1). Aspartyl proteases are a group of proteolytic enzymes including the pepsin family that share the same catalytic apparatus and usually function in acidic conditions (Rao et al., 1998). This fact limits the function of this class of proteases to specific locations in the cell. Aspartyl proteases are ubiquitous in nature and are involved in a myriad of biochemical processes. Well-known aspartyl proteases include pepsin and renin in humans (Davies, 1990). Aspartyl proteases are directly dependent on aspartic acid residues for their catalytic activity and comprise three sub-families: i) family A1, related to pepsin; ii) family A2, retropepsins, and iii) family A3, retropepsins-like. In the A1 family (clan AA) the catalytic Asp residue occurs within the motif Asp-Ser/Thr-Gly. This family contains many secreted enzymes, which are probably synthesized as propeptides with signal peptides (Dash et al., 2003). The aspartyl protease-deduced primary sequences of the P. brasiliensis were analyzed for the presence of the characteristic motif (Asp-Ser/Thr-Gly). Only one protease of this class presents this motif as shown in Table 1. The excerpt of the alignment of this P. brasiliensis aspartyl protease with related sequences is shown in Figure 2. The conserved catalytic motif of known aspartyl proteases and its active site are shown, which are conserved among the compared sequences (Figure 2). Since only the sequence (GenBank accession number AY278218) encoding the homolog of a secreted aspartyl protease of P. brasiliensis was completely sequenced and characterized, it is possible that complete sequencing of other aspartyl proteases found in the P. brasiliensis transcriptome would reveal the expected catalytic triad.
Metalloproteases of Paracoccidioides brasiliensis Zinc-containing metalloproteases are widely distributed in prokaryotic and eukaryotic organisms and are classified into four groups comprehending DD-carboxypeptidases, carboxypeptidases, zincins, and inverzincins (Miyoshi and Shinoda, 2000). One of the most prominent group comprehends the proteins possessing the HEXXH zinc-binding motif, belonging to the zincins superfamily (Miyoshi and Shinoda, 2000). From the eight identified energy-independent zinc metalloproteases in the P. brasiliensis transcriptome, four presented the consensus motif HEXXH which define those proteases as members of the zincins family, as shown in Table 1. The production of such proteases by P. brasiliensis should be of special note, since evidence has been presented identifying the zincins as pathogenic factors in other microorganisms (Klimpel et al., 1994; Matthews et al., 1998). The motif present in carboxypeptidases (HXXE) was also found in the predicted metalloproteases of P. brasiliensis (Table 1). Cysteine proteases of Paracoccidioides brasiliensis Cysteine proteases occur in both prokaryotes and eukaryotes. About 20 subfamilies of cysteine proteases have been recognized. A detailed analysis of the P. brasiliensis transcriptome database reveled two ESTs encoding cysteine proteases ATP independent (Figure 1, Table 1). Active site residues Q, C, H, N obtained by experimental evidence are described in cysteine families in MEROPS database (http://merops.sanger.ac.uk). Two residues (Q and C) were found in one ORF encoding an aminopeptidase of the cysteine family of P. brasiliensis (Table 1). An excerpt of the alignment of this EST with related sequences is shown in Figure 3 and presents those conserved active site residues, which were present in all considered homologues. Among the cysteine proteases, a caspase homolog was detected (Table 1) suggesting that the programmed cell death in P. brasiliensis in its initiation and execution phases could be proteolytically regulated by this class of molecules, as described in other systems (Shi, 2002).
Serine proteases of Paracoccidioides brasiliensis Serine proteases are a family of enzymes that utilize a uniquely activated serine residue in the substrate-binding site to catalytically hydrolyze peptide bonds (Schultz and Liebman, 1997). They are numerous and widespread among virus, bacteria and eukaryotes, suggesting that they are vital to the organisms. Owing to the expanding roles for serine proteases, including a diverse array of physiological functions (Henderson et al., 1992; Froelich et al., 1993), there has been increasing interest in the identification, structural and functional characterization of members of this family. In terms of absolute numbers we identified two energy independent serine proteases in the P. brasiliensis transcriptome (Figure 1, Table 1). The essential amino acid residues forming the catalytic triad (DHS) were detected in one of the deduced ORFs encoding the serine protease of P. brasiliensis (GenBank accession number AY319300). Figure 4 presents the alignment of the deduced amino acid sequence encoding this serine protease, family S08A of P. brasiliensis with related sequences present in MEROPS database. The catalytic triad is conserved among the sequences. Among the identified serine proteases a Kex2 endoprotease was identified (Table 1). The Kex2 endoprotease presented the highest identity to the Kex2 gene of P. brasiliensis described elsewhere (Venancio et al., 2002).
Energy-dependent protease homologues in Paracoccidioides brasiliensis Lon protease The first ATP-dependent protease to be identified in P. brasiliensis was the Lon protease (Barros and Puccia, 2001). The lon gene product is a protein of 1063 amino acids, which presents a single ATP-binding consensus and a serine catalytic site (KDGPSAG). Lon is an endoprotease, cleaving substrates at multiple sites only in the presence of ATP, in several organisms. The protease activity of P. brasiliensis Lon has to be determined. Clps proteases Clps has been described in P. brasiliensis (Table 2). The first Clp protein to be described in the fungus was the ClpB (Jesuino et al., 2002). The ClpB protein of P. brasiliensis presents two ATP-binding domains which places the protein in the class I Clp/HSP100 family (Schirmer et al., 1996). ClpB is also a heat shock protein which is induced upon the mycelium to yeast transition in P. brasiliensis. Heat shock element motifs in the clpB gene promoter region, in addition to the preferential protein expression in yeast cells could suggest a role of ClpB during the temperature upshift that characterizes the infective process by P. brasiliensis. ClpA is another member of the Clp family described in P. brasiliensis (GenBank accession number AY229978) (Table 2). ClpA is a member of proteins that includes the yeast HSP104, which is required for acute thermotolerance (Parsell et al., 1991). The cDNA sequence, which encodes a predicted protein of 927 amino acids, was obtained (Oliveira et al., 2005). The characteristic two nucleotide-binding domains were present in the deduced protein. Other members of the Clp protease family were obtained by analysis of the P. brasiliensis transcriptome. In agreement to Lon, ClpB and ClpA belong to the serine protease family (Table 2). Subunits of unclassified Clps were also obtained. Another member of the AAA superfamily of protease was identified (Table 2) and includes a predicted mitochondrial product, which should be involved in mitochondrial biogenesis. This mitochondrial AAA metalloprotease contains the zinc-binding domain (HEXXH) and could be encoded by a small gene family, as described in other fungi (Shah et al., 2000). Other mitochondrial peptidases were found and listed in Table 2, including members of the inverzincins and DD-carboxypeptidases of the zinc metalloprotease family (Miyoshi and Shinoda, 2000).
Ubiquitin system for protein tagging The ubiquitin system is a highly complex enzymatic system that covalently modifies selected proteins by attachment to the 8-kDa protein ubiquitin. Selective ubiquitin-mediated proteolysis is the dominant mechanism of degradation of cytosolic and nuclear proteins in eukaryotic cells (Finley and Chau, 1991). In this process, a protein substrate is tagged with a poly-ubiquitin chain that mediates interaction with and degradation by the proteasome (Pickart, 2000). Proteasomes The proteasome is the central protease in non-lysosomal ubiquitin-dependent protein degradation, and is involved in protein quality control, antigen processing, signal transduction, cell cycle control, cell differentiation, and apoptosis (Voges et al., 1999). The 26S proteasome is a large protein machine, which is found in both, nucleus and cytoplasm. It consists of the 20S proteasome, which forms the proteolytically active core and a regulatory 19S complex (Glickman et al., 1998). High-resolution crystal structures of the 20S proteasome of the yeast Saccharomyces cerevisiae demonstrated that it is composed of 28 protein subunits, which are arranged into four staggered heptameric rings. Each outer ring comprises seven a-type subunits and each inner contains seven b-type subunits (Groll et al., 1997). The 20S proteasome of higher eukaryotes is also composed of seven distinct a and b subunits, respectively (Krüger et al., 2001). The a subunits are inactive whereas the b subunits build up the hydrolytic chamber. From the seven b subunits, only three are proteolytically active and autoproteolytically matured as active threonine proteases (Groll et al., 1997). The proteolytic activities of the complex in yeast and mammalian proteasomes reside in b1, b2, and b5 subunits (Heinemeyer et al., 1997). By using the primary information from the P. brasiliensis transcriptome database we have annotated ESTs encoding all the a subunits (1 to 7) and six homologues to the 20S b subunit (1 to 6) (Table 3). Of special note is the presence of the b subunits 1, 2 and 5 suggesting that the complex is proteolytically active in P. brasiliensis. Recognizing the polyubiquitin proteolytic signal is one of the many tasks of the 19S complex. Studies have shown that four or more ubiquitin composing chains bind the 19S complex (Thrower et al., 2000). Components of the regulatory 19S complex were found in the P. brasiliensis transcriptome in a total of nine different subunits (Table 3).
Deubiquitinating proteases The deubiquitinating enzymes are defined as a group of proteases which play an important role in the regulation of all processes involving ubiquitin from the processing of poly-ubiquitin precursors into ubiquitin monomers to the targeting or salvage of proteasomal substrates. They are grouped into two classes based on the sequence homology: ubiquitin carboxy-terminal hydrolases (UCHs) and ubiquitin processing proteases, also known as ubiquitin-specific proteases (UBPs). The physiological functions of deubiquitinating enzymes have been elucidated. Studies involving a human gene encoding an ubiquitin-specific protease reveal that the overexpression of this gene can result in deubiquitination of a broad spectrum of cellular proteins with a growth inhibitory effect. This result suggests that this protein may play an important role in regulation of cell growth (Gong et al., 2000). In addition, these enzymes are active in regenerating free ubiquitin after proteins have been targeted to the proteasome (Wing, 2003). A large number of genes encode deubiquitinating enzymes suggesting that many of these have highly specific and regulated functions. The proteins contain conserved motifs with critical cysteine in the active sites. In agreement the UCHs and UBPs in P. brasiliensis are cysteine proteases (Table 4). In S. cerevisiae there is one UCH and sixteen UBPs, whereas higher organisms express an expanded group of enzymes (Yan et al., 2000). In the transcriptome of P. brasiliensis were found three ORFs encoding ubiquitin carboxy terminal hydrolases, a high number when compared to S. cerevisiae. One EST encoding an ubiquitin-specific protease was also found in the transcriptome database (Table 4).
CONCLUDING REMARKS - PUTATIVE ROLE OF PROTEASES IN HOST-PATHOGEN INTERACTION IN PARACOCCIDIOIDES BRASILIENSIS The proteases present in different parasites appear to be relevant for several aspects of host-parasite interactions, quite apart from their obvious participation in the other cellular processes. Information about the seemly putative functions and importance of ATP-independent proteases in P. brasiliensis interaction with host was obtained by comparing them to the described homologues in other systems for whom a function was defined. The putative role in host-fungus interaction for these proteins was deduced by generation of mutants deficient in the genes, or asserting their function as potential antigen or vaccine candidates. Aspartyl proteases are secreted by pathogenic species of Candida in vivo, during infection (De Bernardis et al., 1990). More direct evidence of the implication of SAP proteins in virulence has come from studies of constructing strains harboring disruptions in a number of SAP genes. In all cases, mutants showed decreased virulence in an animal model of disseminated candidiasis (Sanglard et al., 1997; Hube et al., 1997). The SAP 2 confers immune protection against systemic candidiasis in immunized mice and has been postulated as a vaccination target (Villanova et al., 2004). Also, members of this protein family have been considered as antigenic markers of disseminated candidiasis (Morrison et al., 2003). In addition, an aspartyl protease is a component of a protective vaccine in coccidioidomycosis (Johnson et al., 2000). Serine proteases are involved in Aspergillus interaction with the host. A vacuolar serine protease is a major allergen of A. fumigatus (Shen et al., 2003). Also the involvement of serine and cysteine protease in the fungus colonization of the host’s lung tissue has been reported (Kogan et al., 2004). Metalloproteases play different roles in the host-parasite infections. A metalloprotease is a surface antigen in Trypanosoma cruzi and has been postulated as a virulence factor (Cuevas et al., 2003). The protein promotes the attachment of the promastigote form to host cell surface receptors and interacts with the complement system contributing to the ability of the amastigote form of Leishmania spp to survive inside the macrophage (Cheng and Chang, 1986; Joshi et al., 2002). The metalloproteases from the human enteropathogenic Vibrio cholerae accelerate the bacterial attachment to intestinal epithelial cells through digestion of the small intestinal mucosa (Ichinose et al., 1994). Of the proteases produced by A. fumigatus, metalloprotease presenting the consensus zinc-binding motif (HEXXH) has been involved in the infection (Markaryan et al., 1994; Jaton-Ogay et al., 1994). In the current study, we provided an overview of proteases in the P. brasiliensis transcriptome. Research will be directed towards identification of all P. brasiliensis proteases and their functional characterization, as well as its presumed role in the infection by P. brasiliensis. Proteases have enormous potential as drug targets. Perhaps the main reason for this is that protein modification by proteolytic enzymes is such a ubiquitous biological phenomenon that it is difficult to find pathways in which it does not play a part. In the area of proteases of pathogens the potential as drug targets is a great promise because it should be possible to exploit the differences between enzymes of the pathogen and host to produce effective drugs. ACKNOWLEDGMENTS Research supported by MCT/CNPq, CNPq, CAPES, FUB, and UFG. We are thankful to Hugo Costa Paes for English revision. REFERENCES Barrett, A.J., Rawlings, N.D. and Woessner, J.F. (Editors) (1998). Handbook of Proteolytic Enzymes. Academic Press Inc., London, England. Barros, T.F. and Puccia, R. (2001). Cloning and characterization of a LON gene homologue from the human pathogen Paracoccidioides brasiliensis. Yeast 18: 981-988. Carmona, A.K., Puccia, R., Oliveira, M.C., Rodrigues, E.G., Juliano, L. and Travassos, L.R. (1995). Characterization of an exocellular serine-thiol protease activity in Paracoccidioides brasiliensis. Biochem. J. 309: 209-214. Cheng, C.S. and Chang, K.P. (1986). Monoclonal antibody affinity purification of Leishmania membrane glycoprotein and its inhibition of Leishmania-macrophage binding. Proc. Natl. Acad. Sci. USA 83: 100-104. Cuevas, I.C., Cazzulo, J.J. and Sanchez, D.O. (2003). gp63 homologous in Trypanosoma cruzi: surface antigens with metalloprotease activity and a possible role in host cell infection. Infect. Immun. 71: 5739-5749. Dash, C., Kulkarni, A., Dunn, B. and Rao, M. (2003). Aspartyl peptidase inhibitors: Implications in drug development. Crit. Rev. Biochem. Mol. Biol. 38: 89-119. Davies, D.R. (1990). The structure and function of the aspartyl proteases. Annu. Rev. Biophys. Biophys. Chem. 19: 189-215. De Bernardis, F., Agatensi, L., Ross, I.K., Emerson, G.W., Lorenzini, R., Sullivan, P.A. and Cassone, A. (1990). Evidence for a role for secreted aspartate proteinase of Candida albicans in vulvovaginal candidiasis. J. Infect. Dis. 161: 1276-1283. Finley, D. and Chau, V. (1991). Ubiquitination. Ann. Rev. Cell Biol. 7: 25-69. Froelich, C.J., Zhang, X., Turbov, J., Hudeg, D., Winkler, U. and Hanna, W.L. (1993). Human granzyme B degrades aggrecan proteoglycan in matrix synthesized by chondrocytes. J. Immunol. 151: 7161-7171. Glickman, M.H., Rubin, D.M., Fried, V.A. and Finley, D.A. (1998). The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 18: 3149-3162. Gong, L., Kamitani, T., Millas, S. and Yeh, E.T.H. (2000). Identification of a novel isopeptidase with dual specificity for ubiquitin- and NEDD8-conjugated proteins. J. Biol. Chem. 275: 14212-14216. Groll, M., Ditzel, L., Löwe, J., Stock, D., Bochther, M., Bartunkik, H.D. and Huber, R. (1997). Structure of the 20S proteasome from yeast at 2.4 A resolution. Nature 386: 463-471. Heinemeyer, W., Fischer, M., Krimmer, T., Tachon, U. and Wolf, D.H. (1997). The active sites of the eukaryotic 20S proteasome and their involvement in subunit precursor processing. J. Biol. Chem. 272: 25200-25209. Henderson, B.R., Tansey, W.P., Phillips, S.M., Ramshaw, J.A. and Kifford, R.F. (1992). Transcriptional and posttranscriptional activation of urokinase plasminogen activator gene expression in metastatic tumor cells. Cancer Res. 52: 2489-2496. Hube, B. (2000). Exocellular proteases of human pathogenic fungi. Contrib. Microbiol. 5: 126-137. Hube, B., Sanglard, D., Odds, F.C., Hess, D., Monod, M., Schaffer, W., Brown, A.J.P. and Gow, N.A.R. (1997). Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2 and SAP3 of Candida albicans attenuates virulence. Infect. Immun. 65: 3529-3538. Ichinose, Y., Ehara, M., Honda, T. and Mikatani, T. (1994). The effect on enterotoxicity of protease purified from Vibrio cholerae O1. FEMS Microbiol. Lett. 115: 265-271. Jaton-Ogay, K., Paris, S., Huerre, M., Quadroni, M., Falchetto, R., Togni, G., Latge, J.P. and Monod, M. (1994). Cloning and distribution of the gene encoding an extracellular metalloprotease of Aspergillus fumigatus. Mol. Microbiol. 14: 917-928. Jesuíno, R.S.A., Azevedo, M.O., Felipe, M.S.S., Pereira, M. and Soares, C.M.A. (2002). Characterization of a chaperone ClpB homologue of Paracoccidioides brasiliensis. Yeast 19: 963-972. Johnson, S.M., Kerekes, K.M., Zimmermann, C.R., Williams, R.H. and Pappagianis, D. (2000). Identification and cloning of an aspartyl protease from Coccidioides immitis. Gene 241: 213-222. Joshi, P.B., Kelly, B.L., Kamhawi, S., Sacks, D.L. and McMaster, W.R. (2002). Targeted gene deletion in Leishmania major identifies leishmanolysin (GP63) as a virulence factor. Mol. Biochem. Parasitol. 120: 33-40. Klimpel, K.R., Arora, N. and Leppla, S.H. (1994). Anthrax toxin lethal factor contains a zinc metalloprotease consensus sequence which is required for lethal toxin activity. Mol. Microbiol. 13: 1093-1100. Kogan, T.V., Jadoun, J., Mittelman, L., Hirschberg, K. and Osherov, N. (2004). Involvement of secreted Aspergillus fumigatus proteases in disruption of the actin fiber cytoskeleton and loss of focal adhesion sites in infected A549 lung pneumocytes. J. Infect. Dis. 189: 1965-1973. Krüger, E., Kloetzel, P.M. and Enenkel, C. (2001). 20S proteasome biogenesis. Biochimie 83: 289-293. Markaryan, A., Morozova, I., Yu, H. and Kolattukudy, P.E. (1994). Purification and characterization of an elastinolytic metalloprotease from Aspergillus fumigatus and immunoelectron microscopic evidence of secretion of this enzyme by the fungus invading the murine lung. Infect. Immun. 62: 2149-2157. Matthews, R.C., Maresca, B., Burnie, J.P., Cardona, A., Carratu, L., Conti, S., Deepe, G.S., Florez, A.M., Franceschelli, S., Garcia, E., Gargano, L.S., Kobayashi, G.S., McEwen, J.G., Ortiz, B.L., Oviedo, A.M., Polonelli, L., Ponton, J., Restrepos, A. and Storlazzi, A. (1998). Stress proteins in fungal diseases. Med. Mycol. 1: 45-51. McEwen, J.G., Garcia, A.M., Ortiz, B.L., Botero, S. and Restrepo, A. (1995). In search of the natural habitat of Paracoccidioides brasiliensis. Arch. Med. Res. 26: 305-306. Menon, A.S. and Goldberg, A.L. (1987). Protein substrates activate the ATP-dependent protease La by promoting nucleotide binding and release of bound ADP. J. Biol. Chem. 262: 14929-14934. Miyoshi, S. and Shinoda, S. (2000). Microbial metalloproteases and pathogenesis. Microbes Infect. 2: 91-98. Morrison, C.J., Hurst, S.F. and Reiss, E. (2003). Competitive binding inhibition enzyme-linked immunosorbent assay that uses the secreted aspartyl protease of Candida albicans as an antigenic marker for diagnosis of disseminated Candidiasis. Clin. Diagn. Lab. Immunol. 10: 835-848. North, M. (1982). Comparative biochemistry of the proteases of eukaryotic microorganisms. Microbiol. Rev. 46: 308-340. Oliveira, J.C., Castro, N.S., Felipe, M.S.S., Pereira, M. and Soares, C.M.A. (2005). Comparative analysis of the cDNA encoding a ClpA homologue of Paracoccidioides brasiliensis. Mycol. Res. 109: 707-716 [Published online: June 14, 2005]. Parsell, D.A., Sanches, Y., Stitzel, J.D. and Lindquist, S. (1991). Hsp104 is a highly conserved protein with two essential nucleotide binding sites. Nature 353: 270-273. Pickart, C.M. (2000). Ubiquitin in chains. Trends Biochem. Sci. 25: 544-548. Puccia, R., Juliano, M.A., Juliano, L., Travassos, L.R. and Carmona, A.K. (1999). Detection of the basement membrane-degrading proteolytic activity of Paracoccidioides brasiliensis after SDS-PAGE using agarose overlays containing Abz-MKALTLQ-EDDnp. Braz. J. Med. Biol. Res. 32: 645-649. Rao, M.B., Tanksale, A.M., Ghatge, M.S. and Deshpande, V.V. (1998). Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 62: 597-635. Rawlings, N.D. and Barrett, A.J. (1993). Evolutionary families of peptidases. Biochem. J. 290: 205-218. Sanglard, D., Hube, B., Monod, M., Odds, F.C. and Gow, N.A.R. (1997). A triple deletion of the secreted aspartyl protease genes SAP 4, SAP 5 and SAP 6 of Candida albicans causes attenuated virulence. Infect. Immun. 65: 3539-3546. Schirmer, E.C., Glover, J.R., Singer, M.A. and Lindquist, S. (1996). HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21: 289-295. Schultz, R.M. and Liebman, M.N. (1997). Strucutre-function relationship in protein families. In: Textbook of Biochemistry with Clinical Correlations (Devlin, T.M., ed.). 4th edn. Wiley-Liss, New York, NY, USA, 1-116. Shah, Z.H., Hakkaart, G.A., Arku, B., de Jong, L., van der Spek, H., Grivell, L.A. and Jacobs, H.T. (2000). The human homologue of the yeast mitochondrial AAA metalloprotease Yme1p complements a yeast yme1 disruptant. FEBS Lett. 478: 267-270. Shen, H.D., Chou, H., Tam, M.F., Chang, C.Y., Lai, H. and Wang, S.R. (2003). Molecular and immunological characterization of Pen ch18, the vacuolar serine protease major allergen of Penicillium chrysogenuym. Allergy 58: 993-1002. Shi, Y. (2002). Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9: 459-470. Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. and Higgins, D.G. (1997). The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876-4882. Thrower, J.S., Hoffman, S., Rechsteiner, M. and Pckart, C.M. (2000). Recognition of the polyubiquitin preolytic signal. EMBO J. 19: 94-102. Venancio, E.J., Daher, B.S., Andrade, R.V., Soares, C.M., Pereira, I.S. and Felipe, M.S. (2002). The Kex2 gene from the dimorphic and human pathogenic fungus Paracoccidioides brasiliensis. Yeast 19: 1221-1231. Villanova, M., Teixeira, L., Caramalho, I., Torrado, E., Marques, A., Madureira, P., Ribeiro, A., Ferreira, P., Gama, M. and Demengeot, J. (2004). Protection against systemic Candidiasis in mice immunized with secreted aspartyl protease. J. Immunol. 111: 334-342. Voges, D., Zwicki, P. and Baumeister, W. (1999). The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68: 1015-1068. Watson, R.R. (1976). Substrate specificities of aminopeptidases: a specific method for microbial differentiation. Methods Microbiol. 9: 1-14. Wing, S.S. (2003). Deubiquitinating enzymes - the importance of driving in reverse along the ubiquitin-proteasome pathway. Int. J. Biochem. Cell Biol. 35: 590-605. Yan, N., Doelling, J.H., Falbel, T.G., Durski, A.M. and Vierstra, R.D. (2000). The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 124: 1828-1843. Yang, Y.L. (2003). Virulence factors of Candida species. J. Microbiol. Immunol. Infect. 36: 223-228. |
|