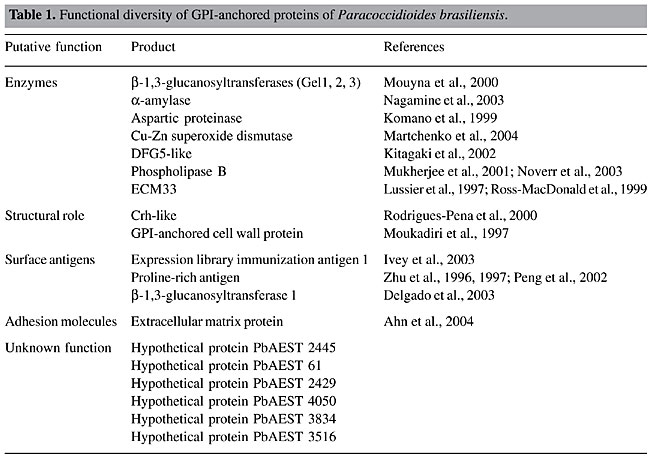
ABSTRACT. Open reading frames in the transcriptome of Paracoccidioides brasiliensis were screened for potential glycosylphosphatidylinositol (GPI)-anchored proteins, which are a functionally and structurally diverse family of post-translationally modified molecules found in a variety of eukaryotic cells. Numerous studies have demonstrated that various GPI anchor sequences can affect the localization of these proteins in the plasma membrane or the cell wall. The GPI anchor core is produced in the endoplasmic reticulum by sequential addition of monosaccharides and phospho-ethanolamine to phosphatidylinositol. The complete GPI anchor is post-translationally attached to the protein carboxyl-terminus by GPI transamidases. Removal of this GPI lipid moiety by phospholipases generates a soluble form of the protein. The identification of putative GPI-attached proteins in the P. brasiliensis transcriptome was based on the following criteria: the presence of an N-terminal signal peptide for secretion and a hydrophobic region in the C-terminus presenting the GPI-attachment site. The proteins that were identified were in several functional categories: i) eight proteins were predicted to be enzymes (Gel1, Gel2, Gel3, a-amylase, aspartic proteinase, Cu-Zn SOD, DFG5, PLB); ii) Ag2/PRA, ELI-Ag1 and Gel1 are probably surface antigens; iii) Crh-like and the GPI-anchored cell wall protein have a putative structural role; iv) ECM33 and Gels (1, 2 and 3) are possibly involved in cell wall biosynthesis, and v) extracellular matrix protein is considered to be an adhesion protein. In addition, eight deduced proteins were predicted to localize in the plasma membrane and six in the cell wall. We also identified proteins involved in the synthesis, attachment and cleaving of the GPI anchor in the P. brasiliensis transcriptome. Key words: Paracoccidioides brasiliensis, GPI-anchored proteins, Plasma membrane, Cell wall INTRODUCTION Cell surface membrane proteins constitute an important class of biomolecules in living cells, as they are at the interface with the surrounding environment. Most eukaryote membrane proteins are post-translationally modified, and a subset of them is modified by the attachment of a glycosylphosphatidylinositol (GPI) moiety at the C-terminal end of the protein (Ferguson et al., 1988). Although fungal and mammalian cells contain the same mechanism by which they attach carbohydrates to nascent proteins, mammalian GPI anchors tether proteins to cell membranes, whereas in fungal cells GPI anchors are also used to covalently link proteins to cell wall glucans (Varki et al., 1999). GPI-modified proteins are widely found in lower and higher eukaryotes (Eisenhaber et al., 2001). The primary sequence of GPI proteins share a general pattern, with N-terminal signal peptides and C-terminal features that mediate GPI anchor addition at an amino acid residue designated the omega (w)-site (Hamada et al., 1998b). GPI anchor addition occurs in the endoplasmic reticulum (ER), following proteolytic cleavage of the C-terminal propeptide (Orlean, 1997). In addition to these signal sequences, the GPI proteins present a serine-threonine-rich sequence that provides sites for glycosylation. Moreover, the cellular localization of GPI-anchored proteins seems to be at least partly determined by basic or hydrophobic residues in the w-region (Caro et al., 1997; Vossen et al., 1997; Hamada et al., 1998b, 1999). The core structure of the GPI anchor consists of a single phospholipid spanning the membrane and a head group consisting of a phosphodiester-linked inositol, to which a glucosamine is linked, a linear chain of three mannose sugars linked to glucosamine and an ethanolamine phosphate (EtNP) linked to the terminal mannose. Composition differences in the lipid portion and side chain substitutions in the tetrasaccharide backbone of the conserved head-group promote variants in the structure of the GPI anchor. One of the most prominent aspects of GPI anchor diversity is glycan substitution of the conserved mannose residues (McConville and Ferguson, 1993). The biosynthesis of the GPI moiety occurs in the ER, and the complete GPI anchor is fully assembled prior to attachment to the protein. A series of sequential enzymatic steps adds the various GPI components. GPI proteins enter the ER where the GPI anchor is covalently added to the w-site by a transamidase complex of at least five proteins (Fraering et al., 2001; Hong et al., 2003). The GPI-anchored proteins are transported from the ER to the Golgi apparatus in distinct vesicles from the non-GPI-anchored proteins (Muniz et al., 2001). A Rab GTPase is specifically required for GPI protein trafficking. Also, the tethering factors Vso1 and Sec34/35p are necessary for the sorting of GPI-anchored proteins upon ER exit (Morsomme and Riezman, 2002). Most available evidence suggests that there are two terminal fates for GPI proteins. They can reside at the plasma membrane (GPI-anchored plasma membrane proteins) or reside at the cell wall (Lu et al., 1994). Caro et al. (1997) proposed, based on in silico analysis of GPI-anchored proteins of Saccharomyces cerevisiae, that a signal of two basic amino acids in the four residues upstream to the w-site acts to retain the protein at the plasma membrane. Hamada et al. (1998b, 1999) suggested that in the absence of this retention signal, hydrophobic amino acids at positions 2, 4, and 5, upstream to the w-site act positively to localize the protein to the cell wall. The intact GPI anchor confers an amphiphilic character to the protein, which by the action of phospholipases (PLs) cleaving the ester bond of the phosphatidylinositol (PI), render the protein hydrophilic. In this way, a proposed role for the GPI anchor and their solubilizing PLs is that it may be an alternative to proteolysis for the regulated release of proteins from membranes (Ehlers and Riordan, 1991). The location of GPI proteins makes them ideal candidates for such function. Several studies have now established that GPI-anchored proteins are a large class of functionally diverse proteins. They can be enzymes, surface antigens, adhesion molecules, or surface receptors (Chatterjee and Mayor, 2001; Hoyer, 2001; Sundstrom, 2002). GPI-anchored proteins reported in various microbial pathogens have been shown to be immunogenic and are suggested to be important virulence factors (Hung et al., 2002; McGwire et al., 2002). In addition, GPI-bound proteins can display enzymatic properties, playing an active role in cell wall biosynthesis (Hartland et al., 1996; Mouyna et al., 2000). In fungi, synthesis of GPI anchors is essential for viability, since their cell wall mannoproteins require a GPI anchor so that they can be covalently incorporated into the cell wall (Leidich et al., 1994). Yeast has been extensively used to study the GPI-anchoring system, and it is now well understood (Ash et al., 1995; van der Vaart et al., 1995). However, in contrast to the case for S. cerevisiae, little is known about the structure and biosynthesis of the GPI anchor in filamentous fungi. Aspergillus fumigatus presents about nine GPI-anchored protein homologs to the yeast counterparts (Bruneau et al., 2001). Fontaine et al. (2003) characterized four GPI-anchored proteins from a membrane preparation of A. fumigatus. In contrast to yeast, only ceramide was found in the GPI anchor structure of A. fumigatus. The glycan moiety is mainly a linear pentomannose structure, linked to a glucosamine residue. The thermal dimorphic fungus Paracoccidioides brasiliensis causes paracoccidioidomycosis, the leading endemic deep mycosis in Latin America. The disease may develop as different forms, ranging from benign and localized to severe and disseminated forms (Franco et al., 1993). Fungal conidia start the infection, which undergo conversion to the yeast parasitic phase in human lungs (McEwen et al., 1987). The morphological switch from mycelia to yeasts is the most important biological feature that enables P. brasiliensis to colonize, invade and survive in host tissues during infection (San-Blas et al., 2002). Previous reports described that P. brasiliensis makes use of GPI as a means of membrane anchorage of surface proteins (Heise et al., 1995). The addition of complete GPI anchors is required for morphogenesis, virulence and for host-fungus interactions (Richard et al., 2002; Sundstrom, 2002; Delgado et al., 2003). These reasons can be invoked to account for the importance of GPI-anchored proteins in P. brasiliensis. An efficient method for retrieving novel GPI proteins is a genome sequence-based approach. Computational methods provide a useful starting point for genome-wide screening of potential GPIs in a variety of organisms. Saccharomyces cerevisiae DNA sequencing and Von Heijine algorithm studies identified 58 potential GPI-anchored proteins (Caro et al., 1997). Recently, P. brasiliensis transcriptome information (https://www.biomol.unb.br/Pb) have been obtained and released in public databases. The availability of this transcriptome gives us a new strategy for identifying genes that are likely GPI proteins. We report 20 putative GPI-anchored predicted proteins in the P. brasiliensis transcriptome. MATERIAL AND METHODS Sequence and motif similarity is the most commonly used method for assigning a putative function to newly discovered genes. The identification of putative GPI-anchored proteins was based on the following criteria: i) the presence of an N-terminal signal peptide for secretion; ii) a hydrophobic tail, and iii) the GPI-attachment site. Two GPI-anchored prediction tools, big-PI fungal predictor (http://mendel.imp.univie.ac.at/gpi/fungi/gpi_fungi.html) (Eisenhaber et al., 2004) and DGPI (http://129.194.185.165/dgpi/index_en.html) were used to screen the P. brasiliensis GPI-anchored proteins. The presence of a signal sequence for import into ER was confirmed by using SignalP version 3.0 (http://www.cbs.dtu.dk/services/SignalP/) (Nielsen et al., 1997; Bendtsen et al., 2004). The presence of hydrophobic regions was analyzed with DAS (http://www.sbc.su.se/~miklos/DAS/) (Cserzo et al., 1997) and PSORT II (http://www.psort.org/) (Horton and Nakai, 1997). PSORT II was also used for protein localization predictions. BLAST searches were performed at NCBI (http://www.ncbi.nlm.nih.gov/BLAST/) (Altschul et al., 1997) and Pfam (http://www.sanger.ac.uk/software/pfam/index.shtml) (Bateman et al., 2002). A phylogenetic tree was constructed by multiple sequence alignments by using the Clustal X program, version 1.8 and the neighbor joining method (Thompson et al., 1997). Robustness of branches was estimated using 100-boot-strapped replicates. The amino acid sequences were visualized using the TreeView software. RESULTS AND DISCUSSION Putative GPI-anchored proteins of P. brasiliensis Several studies have now established that GPI-anchored proteins are a large class of functionally diverse proteins. The predicted GPI-anchored proteins of P. brasiliensis could be enzymes, surface antigens, or adhesion molecules, and they have a structural role in the cell wall biogenesis (Table 1). For instance, a-amylase, proline-rich antigen/antigen 2 (PRA/Ag2), Cu-Zn superoxide dismutase (Cu-Zn SOD), GPI-anchored cell wall, ECM33, Crh-like, DFG5-like, PLB, extracellular matriz protein (EMP), aspartic proteinase precursor, expression library immunization antigen 1 (ELI Ag1) and b-1,3-glucanosyltransferase (Gels 1, 2 and 3) proteins were found. Their predicted functions were obtained by comparison to the homologs for which a role has been defined.
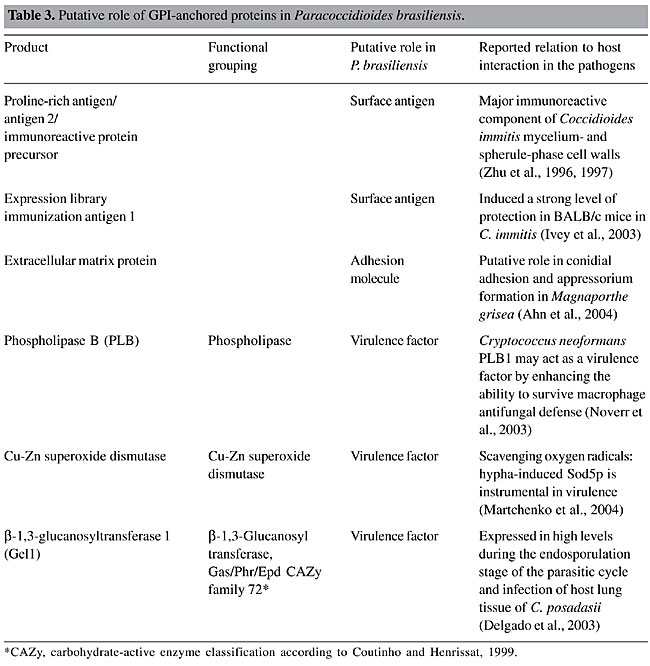
In our 20 predicted GPI-anchored proteins nine in our list are supposed to have enzymatic activity. The a-amylase enzyme is located on the cell wall of fungi, and it plays a crucial role in the fermentation process in yeast (Yabuki and Fukui, 1970; Nagamine et al., 2003). Aspartic proteinase could act in the processing of cell wall precursors or precursors of enzymes involved in cell wall synthesis or remodeling (Komano et al., 1999). Eukaryotic Cu- and Zn-containing superoxide dismutase 1 (SOD1) is a key superoxide scavenging enzyme that is largely localized in the cytosol but is also found in the intermembrane space of mitochondria and in other organelles (Weisiger and Fridovich, 1973; Chang et al., 1988; Keller et al., 1991; Okado-Matsumoto and Fridovich, 2001; Sturtz et al., 2001). Some of the newly identified proteins, ECM33 and DFG5-like, have been reported to be involved in cell wall biogenesis (Lussier et al., 1997; Ross-MacDonald et al., 1999; Kitagaki et al., 2002) and cell growth at high temperature (Terashima et al., 2003). In addition, the Gel family is also required for proper cell wall assembly and morphogenesis due to their activity elongating b-1,3-glucans of human fungal pathogens (Mouyna et al., 2000). Two proteins have been reported to have a structural role: Crh-like, which has a putative glycosidase domain and could be involved in the development of cell wall architecture (Rodriguez-Pena et al., 2000), and GPI-anchored cell wall protein, which has a structural role in association with the glucan network, since both have the same localization (Moukadiri et al., 1997). All known GPI-anchored proteins share a number of common features, including the predominantly hydrophobic region in the C-terminus, which most likely functions as a recognition signal for a transamidase system, the absence of transmembrane domains in the mature molecule and the presence of a cleavable N-terminal secretion signal for translocation into the ER. Based on the algorithms described above, we detected 20 predicted GPI-anchored proteins in the P. brasiliensis transcriptome (Table 2). In mammalian cells, over 100 cell surface proteins are putative GPI-anchored proteins (Low, 1989; Kinoshita et al., 1995). Fifty-eight potential GPI-anchored proteins were identified in the S. cerevisiae genome (Caro et al., 1997). Among the identified GPI proteins, 16 presented the N-terminal signal peptides (Table 2). Among the 20 P. brasiliensis GPI-anchored proteins we were able to detect C-terminal regions in 11 predicted proteins (Table 2). Several residues of S and T, potential sites for O-glycosylation, were detected, even in the partial sequences (Table 2). GPI proteins usually have a high percentage of S and T residues, the side-chains of which are potential sites for O-glycosylation (Klis et al., 2002). The S/T content in the putative P. brasiliensis GPI proteins varies from 9 to 28%, with an average of 20%, which is similar to predicted GPI-anchored proteins of Neurospora crassa (21%) and slightly lower than in S. cerevisiae (25%), Candida albicans (28%) and Schizosaccharomyces pombe (29%) (de Groot et al., 2003).      Putative cellular localization of the predicted GPI-anchored proteins of P. brasiliensis Although most GPI-anchored proteins in yeast and other fungi localize to the cell wall, some are believed to reside at the plasma membrane. Evidence indicates that the amino acids immediately upstream to the w-site serve as the signal determining protein localization. Two kinds of signals have been proposed for GPI-anchored protein cellular localization: i) dibasic residues (K and/or R) in a short w-minus region are favored in proteins that are predominantly localized in the plasma membrane (Caro et al., 1997; Vossen et al., 1997) and ii) the specific amino acid residues V, I or L 4 or 5 amino acids upstream of the GPI-attachment site (the w-site) and Y or N at the w-2 site have been shown to act as a positive signal for cell wall localization (Hamada et al., 1998b, 1999). In order to predict the cellular localization of putative GPI-anchored proteins of the P. brasiliensis transcriptome, we analyzed the corresponding amino acids in the w-minus region and also examined the results of k-NN prediction (PSORT II server) (data not shown). We also compared those analyses to data from other organisms. Accordingly, among the 20 GPI-anchored proteins, 11 sequences which presented at least the 42 last amino acids in the C-terminal region were selected to study their putative localization (Table 2). The three proteins of the Gel family, the ECM33 protein and the hypothetical protein PbAEST 2429 presented basic motifs upstream to the predicted w-site, as detected by the big-PI fungal predictor (Eisenhaber et al., 2004). These results were compatible with the k-NN prediction (Horton and Nakai, 1997) and with the literature description of plasma membrane localization (Vai et al., 1991; Hamada et al., 1998a; Terashima et al., 2003). However, no basic amino acid was found in the w-minus region for the Cu-Zn SOD and DFG5-like proteins. In both, plasma membrane localization prevails, as described by Karpinska et al. (2001), Kitagaki et al. (2002) and Spreghini et al. (2003). The PRA/Ag2 and GPI-anchored cell wall proteins were predicted as putatively anchored in the cell wall on the basis of descriptions from Coccidioides immitis (Zhu et al., 1996) and S. cerevisiae (Moukadiri et al., 1997; Hamada et al., 1998a), respectively. Two hypothetical proteins, PbEST 2445 and PbEST 61, were predicted as cell wall proteins, only by the PSORT II analysis. GPI-anchored proteins putatively associated with the fungus host interaction Recent studies suggest that the GPI proteins are instrumental in fungal adhesion, recognition by host receptors, and may play a role in cell wall expansion. GPI-anchored proteins are leading vaccine candidates that are thought to be of major importance for infection (Smythe et al., 1988; Delgado et al., 2003). Table 3 shows some P. brasiliensis proteins that could be involved in host interaction and virulence. Two of the newly identified proteins, PRA/Ag2 and ELI Ag1, have been reported to be surface antigens. PRA/Ag2 is a component of a glycopeptide, which is probably the main T-cell-reactive component of C. immitis cell walls (Zhu et al., 1996). Also, the recombinant PRA/Ag2 protein is reactive with sera from patients with active coccidioidomycosis (Zhu et al., 1997). This protein is suggested to have an endoglucanase activity and to be important for spherule cell-wall morphogenesis during the infection process by C. immitis (Zhu et al., 1996). It is located in the fungal cell wall (Galgiani et al., 1992), most probable attached to the cell wall matrix (Peng et al., 2002). The expression of this protein can be considered phase specific since it is up-regulated during the spherule phase in C. immitis (Galgiane et al., 1992; Peng et al., 1999). ELI Ag1 is the first protective C. immitis antigen that has been identified by expression library immunization, inducing a strong level of protection in BALB/c mice. The mechanism by which this antigen protects mice against a lethal challenge with C. immitis arthroconidia is not yet known (Ivey et al., 2003).
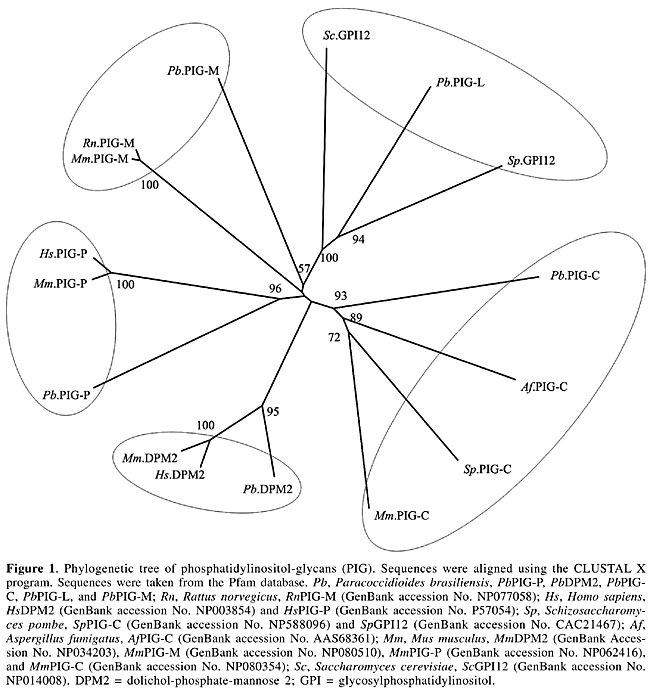
We identified EST homologs to the EMP of Magnaporthe grisea. Although the function of EMP1 remains unclear at the biochemical level, it is suggested that it has a role in sensing a surface signal and/or transmitting a signal into the cell to promote conidial adhesion and appressorium formation in M. grisea (Ahn et al., 2004). Among the identified enzymes, PLB, Cu-Zn SOD and Gel1 are reported as necessary for the virulence of fungal pathogens. It has been postulated that PLs assist in the penetration of phospholipid-rich host barriers, such as membranes and lung surfactant (Cox et al., 2001). Supporting evidence for this role has been shown by deletion of the PLB1 gene in C. albicans, which results in a significant reduction in the ability of the pathogen to traverse the stomach mucosa and disseminate hematogenously to the liver (Mukherjee et al., 2001). Furthermore, PLB1 of Cryptococcus neoformans may act as a virulence factor, by enhancing the ability to survive the macrophage antifungal defenses, possibly by facilitating fungal eicosanoid production during cryptococcal infection (Noverr et al., 2003). The main function of SOD is to scavenge O2- radicals generated in various physiological process, thus preventing the oxidation of biological molecules (Liochev and Fridovich, 1994; Fridovich, 1995). SOD can be classified according to metal co-factor(s) bound to them. Cu-Zn SOD has copper and zinc as metal co-factors (Martchenko et al., 2004). Candida albicans Sod1 was shown to protect cells against extracellular superoxide radicals produced by macrophages, and it was reported to be important for the virulence of C. albicans in a mouse model (Hwang et al., 2002). It was found that mice immunized with the recombinant Gel1 of Coccidioides posadasii and infected against a lethal challenge of this pathogen had a significant reduction in fungal burden and increased survival compared to nonimmune mice. The mature Gel1 was immunolocalized to the surface of endospores, and the highest level of the Gel1 mRNA was detected during the endosporulation stage of the parasitic cycle (Delgado et al., 2003). Furthermore, it was found that two homologous genes in C. albicans are pH-regulated and required for virulence. These genes nominally include PHR1, a gene expressed maximally at pH 5.5 to 8.0, which encodes a protein promoting systemic infection of mice, and a PHR2 gene, the expression pattern of which is the inverse and is involved in pathogenesis in a mouse model of vaginal infection (Saporito-Irwin et al., 1995; Muhlschlegel and Fonzi, 1997; De Bernardis et al., 1998). Phosphatidylinositol-glycan proteins and transamidases The biosynthesis of GPI occurs on the membrane of the ER by the sequential addition of sugar residues to PI by the action of glycosyltransferases (Stevens, 1995). The common core structure of GPI consists of inositol phospholipid, GlcN, three mannoses and EtNP (Ferguson and Williams, 1988). Genes encoding the enzymes in GPI biosynthesis have been identified by cloning, sequencing and by using the techniques of knock out and rescue. In mammals, around 20 genes participate in this pathway and have been identified as phosphatidylinositol-glycan (PIG) gene products (Ferguson, 1999; Kinoshita and Inoue, 2000; McConville and Menon, 2000). The glycosyltransferase complex composed by the proteins PIG-A, PIG-C, PIG-H, GPI1, PIG-P, and DPM2 (dolichol-phosphate-mannose 2) catalyzes the first step in the GPI synthesis (Watanabe et al., 2000). PIG-A encodes a subunit of GPI-N-acetylglucosamine transferase (Mayor and Riezman, 2004). Table 4 summarizes the PIGs found in the P. brasiliensis transcriptome. PIG-H and GPI1 encoding ESTs were not detected in the P. brasiliensis transcriptome. Studies on S. cerevisiae had shown that the Gpi12 homolog of PIG-L participates in the second step of GPI synthesis (Watanabe et al., 1999) and the mannosylation reactions are mediated by PIG-M (GPI-a-1-4 mannosyltransferase) and PIG-B (GPI-a-1-2 mannosyltransferase) (Kinoshita and Inoue, 2000). Only PIG-L was found in the P. brasiliensis transcriptome (Table 4). The EtNP transfer to the first and third mannose residues is mediated by PIG-N and PIG-F and PIG-O, respectively. The first two ESTs had not been detected in our analysis. 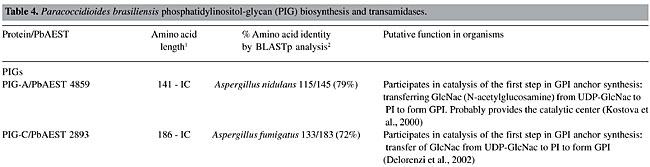    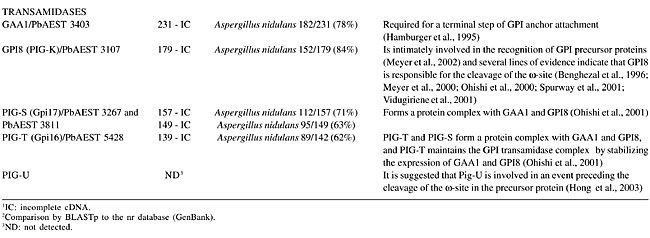 Attachment of the GPI to the protein involves cleavage of the lumenally located pre-protein at a hydrophobic stretch, followed by the attachment of the cleaved sequence to the fully assembled GPI via a transamidase reaction (Udenfriend and Kodukula, 1995). Components of the transamidase complex have been identified in yeast and other organisms (Hamburger et al., 1995; Ohishi et al., 2000). Humans and S. cerevisiae GPI transaminidases are well conserved, containing five homologous components (Hong et al., 2003). Five human components, GAA1 (glycosylphosphatidylinositol anchor attachment 1), GPI8, PIG-S, PIG-T, and PIG-U are homologous to yeast Gaa1p, Gpi8p, Gpi17p, Gpi16p, and Cdc91p, respectively (Fraering et al., 2001; Ohishi et al., 2001). Several lines of evidence indicate that GPI8/Gpi8p are the catalytic components responsible for the cleavage of the GPI-attachment signal sequences (Benghezal et al., 1996; Meyer et al., 2000; Ohishi et al., 2000; Spurway et al., 2001; Vidugiriene et al., 2001). All of those encoding transamidase ESTs were detected in our analysis of the P. brasiliensis transcriptome, with the exception of ESTs encoding PIG-U (Table 4). Phylogenetic relationships of PbPIGs were generated with PIGs available on the Pfam database. The PIGs were well resolved into clades corresponding respectively to PIG-C, PIG-DPM2, PIG-P, PIG-M, and PIG-L (Figure 1). Consequently, we suggest conservation of PIG sequences during evolution.
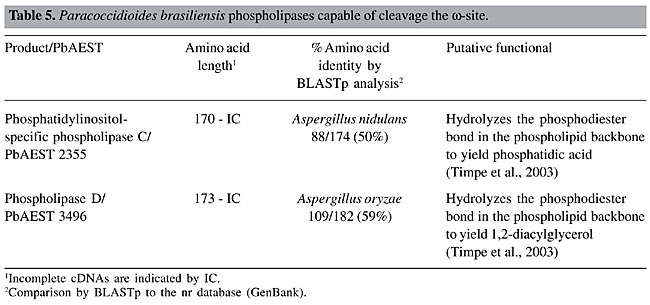
GPI solubilizing phospholipases The intact GPI anchor confers an amphiphilic character to the proteins, which by the action of PLs cleaving the ester bond of the PI, render the protein hydrophilic (Stambuk and Cardoso de Almeida, 1996). Thus, a proposed role for the GPI anchor and their solubilizing PLs is that it may be an alternative to proteolysis for the regulated release of proteins from membranes (Ehlers and Riordan, 1991). The term “phospholipases” refers to a heterogeneous group of enzymes that are able to hydrolyze one or more ester linkages in glycerophospholipids (Cox et al., 2001). The action of PLs can result in the destabilization of membranes, cell lysis and release of lipid second messengers (Schmiel and Miller, 1999; Ghannoum, 2000). Although all PLs target phospholipids as substrates, each enzyme has the ability to cleave a specific ester bond (Cox et al., 2001). Several mammalian PL activities that seem to be capable of removing the GPI anchors from proteins have been reported (Low and Saltiel, 1988). In P. brasiliensis, Heise et al. (1995) reported the detection of a potent PLC capable of selectively hydrolyzing the GPI anchor, with the consequent release of proteins. The search for cDNAs homologous to PLs in the P. brasiliensis transcriptome revealed two open reading frames with high sequence homology to PI-PLC and PLD of A. nidulans and A. oryzae, respectively (Table 5). This finding suggests that PI-PLC and PLD could be capable of hydrolyzing the GPI anchor in P. brasiliensis. The GPI-specific PLC, which is another type of phospholipase C capable of cleaving the GPI anchor, was not found in the P. brasiliensis transcriptome.
CONCLUDING REMARKS The cell wall is a plastic and dynamic structure that is constantly changing in response to environmental signals and to different stages of the fungal cell cycle. GPI anchoring is a eukaryotic mechanism for attaching proteins to the cell surface. In fungi, GPI proteins are known to be either covalently incorporated into the cell wall network or to remain attached to the plasma membrane. The GPI-anchored proteins localized in the cell wall may determine surface hydrophobicity and antigenicity, and they are reported from various microbial pathogens as immunogenic and adhesion molecules; they have also been suggested to be important virulence factors. On the other hand, the GPI proteins localized in the plasma membrane are known to play a role in cell wall biosynthesis and remodeling. This is the first analysis of P. brasiliensis GPI-anchored proteins in the fungus transcriptome. Many of the identified proteins can be broadly categorized as being involved in cell wall remodeling, in host-fungus interaction, providing some insight into the purposes of GPI-anchoring. ACKNOWLEDGMENTS Research supported by MCT/CNPq, CNPq, CAPES, FUB, and UFG. We are thankful to Hugo Costa Paes for English revision. REFERENCES Ahn, N., Kim, S., Choi, W., Im, K.H. and Lee, Y.H. (2004). Extracellular matrix protein gene, EMP1, is required for appressorium formation and pathogenicity of the rice blast fungus, Magnaporthe grisea. Mol. Cell 17: 166-173. Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. Ash, J., Dominguez, M., Bergeron, J.J., Thomas, D.Y. and Bourbonnais, Y. (1995). The yeast proprotein convertase encoded by YAP3 is a glycophosphatidylinositol-anchored protein that localizes to the plasma membrane. J. Biol. Chem. 270: 20847-20854. Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M. and Sonnhammer, E.L. (2002). The Pfam protein families Database. Nucleic Acids Res. 30: 276-280. Bendtsen, J.D., Nielsen, H., von Heijne, G. and Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 40: 783-795. Benghezal, M., Benachour, A., Rusconi, S., Aebi, M. and Conzelmann, A. (1996). Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J. 15: 6575-6583. Bruneau, J.M., Magnin, T., Tagat, E., Legrand, R., Bernard, M., Diaquin, M., Fudali, C. and Latge, J.P. (2001). Proteome analysis of Aspergillus fumigatus identifies glycosylphosphatidylinositol-anchored proteins associated to the cell wall biosynthesis. Electrophoresis 22: 2812-2823. Caro, L.H., Tettelin, H., Vossen, J.H., Ram, A.F., van den Ende, H. and Klis, F.M. (1997). In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13: 1477-1489. Chang, L.Y., Slot, J.W., Geuze, H.J. and Crapo, J.D. (1988). Molecular immunocytochemistry of the CuZn superoxide dismutase in rat hepatocytes. J. Cell Biol. 107: 2169-2179. Chatterjee, S. and Mayor, S. (2001). The GPI-anchor and protein sorting. Cell. Mol. Life Sci. 58: 1969-1987. Coutinho, P.M. and Henrissat, B. (1999). Carbohydrate-active enzymes: an integrated database approach. In: Recent Advances in Carbohydrate Bioengineering (Gilbert, H.J., Davics, G., Henrissat, B. and Svensson, B., eds.). The Royal Society of Chemistry, Cambridge, UK, pp. 3-12. Cox, G.M., McDade, H.C., Chen, S.C., Tucker, S.C., Gottfredsson, M., Wright, L.C., Sorrell, T.C., Leidich, S.D., Casadevall, A., Ghannoum, M.A. and Perfect, J.R. (2001). Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 39: 166-175. Cserzo, M., Wallin, E., Simon, I., von Heijne, G. and Elofsson, A. (1997). Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10: 673-676. De Bernardis, F., Muhlschlegel, F.A., Cassone, A. and Fonzi, W.A. (1998). The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect. Immun. 66: 3317-3325. de Groot, P.W.J., Hellingwerf, K.J. and Klis, F.M. (2003). Genome-wide identification of fungal GPI proteins. Yeast 20: 781-796. Delgado, N., Xue, J., Yu, J.J., Hung, C.Y. and Cole, G.T. (2003). A recombinant beta-1,3-glucanosyltransferase homolog of Coccidioides posadasii protects mice against coccidioidomycosis. Infect. Immun. 71: 3010-3019. Delorenzi, M., Sexton, A., Shams-Eldin, H., Schwarz, R.T., Speed, T. and Schofield, L. (2002). Genes for glycosylphosphatidylinositol toxin biosynthesis in Plasmodium falciparum. Infect. Immun. 70: 4510-4522. Ehlers, M.R.W. and Riordan, J.F. (1991). Membrane proteins with soluble counterparts: role of proteolysis in the release of transmembrane proteins. Biochemistry 30: 10065-10074. Eisenhaber, B., Bork, P. and Eisenhaber, F. (2001). Post-translational GPI lipid anchor modification of proteins in kingdoms of life: analysis of protein sequence data from complete genomes. Protein Eng. 14: 17-25. Eisenhaber, B., Schneider, G., Wildpaner, M. and Eisenhaber, F. (2004). A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Mol. Biol. 337: 243-253. Ferguson, M.A. and Williams, A.F. (1988). Cell-surface anchoring of proteins via glycosyl-phosphatidylinositol structures. Annu. Rev. Biochem. 57: 285-320. Ferguson, M.A., Homans, S.W., Dwek, R.A. and Rademacher, T.W. (1988). Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science 239: 753-759. Ferguson, M.A.J. (1999). The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell. Sci. 112: 2799-2809. Fontaine, T., Magnin, T., Melhert, A., Lamont, D., Latge, J.P. and Ferguson, M.A.J. (2003). Structures of glycosylphosphatidylinositol membrane anchors from Aspergillus fumigatus membrane proteins. Glycobiology 13: 169-177. Fraering, P., Imhof, I., Meyer, V., Strub, J.M., van Dorsselaer, A., Vionnet, C. and Conzelmann, A. (2001). The GPI transamidase complex of Saccharomyces cerevisiae contains Gaa1p, Gpi8p and Gpi16p. Mol. Biol. Cell 12: 3295-3306. Franco, M., Peracoli, M.T., Soares, A., Montenegro, R., Mendes, R.P. and Meira, D.A. (1993). Host-parasite relationship in paracoccidioidomycosis. Curr. Top. Med. Mycol. 5: 115-149. Fridovich, I. (1995). Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 64: 97-112. Galgiani, J.N., Sun, S.H., Dugger, K.O., Ampel, N.M., Grace, G.G., Harrison, J. and Wieden, M.A. (1992). An arthroconidial-spherule antigen of Coccidioides immitis: differential expression during in vitro fungal development and evidence for humoral response in humans after infection or vaccination. Infect. Immun. 60: 2627-2635. Ghannoum, M.A. (2000). Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev. 13: 122-143. Hamada, K., Fukuchi, S., Arisawa, M., Baba, M. and Kitada, K. (1998a). Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol. Gen. Genet. 258: 53-59. Hamada, K., Terashima, H., Arisawa, M. and Kitada, K. (1998b). Amino acid sequence requirement for efficient incorporation of glycosylphosphatidylinositol-associated proteins into the cell wall of Saccharomyces cerevisiae. J. Biol. Chem. 273: 26946-26953. Hamada, K., Terashima, H., Arisawa, M., Yabuki, N. and Kitada, K. (1999). Amino acid residues in the omega-minus region participate in cellular localization of yeast glycosylphosphatidylinositol-attached proteins. J. Bacteriol. 181: 3886-3889. Hamburger, D., Egerton, M. and Riezman, H. (1995). Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J. Cell Biol. 129: 629-639. Hartland, R.P., Fontaine, T., Debeaupuis, J.P., Simenel, C., Delepierre, M. and Latgé, J.P. (1996). A novel beta-(1-3)-glucanosyltransferase from the cell wall of Aspergillus fumigatus. J. Biol. Chem. 271: 26843-26849. Heise, N., Travassos, L.R. and de Almeida, M.L. (1995). Paracoccidioides brasiliensis expresses both glycosylphosphatidylinositol-anchored proteins and a potent phospholipase C. Exp. Mycol. 19: 111-119. Hong, Y., Maeda, Y., Watanabe, R., Ohishi, K., Mishkind, M., Riezman, H. and Kinoshita, T. (1999a). Pig-N, a mammalian homologue of yeast Mcd4p, is involved in transferring phosphoethanolamine to the first mannose of the glycosylphosphatidylinositol. J. Biol. Chem. 274: 35099-35106. Hong, Y., Ohishi, K., Watanabe, R., Endo, Y., Maeda, Y. and Kinoshita, T. (1999b). GPI1 stabilizes an enzyme essential in the first step of glycosylphosphatidylinositol biosynthesis. J. Biol. Chem. 274: 18582-18588. Hong, Y., Maeda, Y., Watanabe, R., Inoue, N., Ohishi, K. and Kinoshita, T. (2000). Requirement of PIG-F and PIG-O for transferring phosphoethanolamine to the third mannose in glycosylphosphatidylinositol. J. Biol. Chem. 275: 20911-20919. Hong, Y., Ohishi, K., Kang, J.Y., Tanaka, S., Inoue, N., Nishimura, J., Maeda, Y. and Kinoshita, T. (2003). Human PIG-U and yeast Cdc91p are the fifth subunit of GPI transamidase that attaches GPI-anchors to proteins. Mol. Biol. Cell 14: 1780-1789. Horton, P. and Nakai, K. (1997). Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5: 147-152. Hoyer, L.L. (2001). The ALS gene family of Candida albicans. Trends Microbiol. 9: 176-180. Hung, C.Y., Yu, J.J., Seshan, K.R., Reichard, U. and Cole, G.T. (2002). A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect. Immun. 70: 3443-3456. Hwang, C.S., Rhie, G.E., Oh, J.H., Huh, W.K., Yim, H.S. and Kang, S.O. (2002). Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology 148: 3705-3713. Ivey, F.D., Magee, D.M., Woitaske, M.D., Johnston, S.A. and Cox, R.A. (2003). Identification of a protective antigen of Coccidioides immitis by expression library immunization. Vaccine 21: 4359-4367. Karpinska, B., Karlsson, M., Schinkel, H., Streller, S., Suss, K.H., Melzer, M. and Wingsle, G. (2001). A novel superoxide dismutase with a high isoelectric point in higher plants. Expression, regulation, and protein localization. Plant Physiol. 126: 1668-1677. Keller, G.A., Warner, T.G., Steimer, K.S. and Hallewell, R.A. (1991). Cu,Zn superoxide dismutase is a peroxisomal enzyme in human fibroblasts and hepatoma cells. Proc. Natl. Acad. Sci. USA 88: 7381-7385. Kinoshita, T. and Inoue, N. (2000). Dissecting and manipulating the pathway for glycosylphosphatidy linositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 4: 632-638. Kinoshita, T., Inoue, N. and Takeda, J. (1995). Defective glycosyl phosphatidylinositol anchor synthesis and paroxysmal nocturnal hemoglobinuria. Adv. Immunol. 60: 57-103. Kitagaki, H., Wu, H., Shimoi, H. and Ito, K. (2002). Two homologous genes, DCW1 (YKL046c) and DFG5, are essential for cell growth and encode glycosylphosphatidylinositol (GPI)-anchored membrane proteins required for cell wall biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 46: 1011-1022. Klis, F.M., Mol, P., Hellingwerf, K. and Brul, S. (2002). Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26: 239-256. Komano, H., Rockwell, N., Wang, G.T., Krafft, G.A. and Fuller, R.S. (1999). Purification and characterization of the yeast glycosylphosphatidylinositol-anchored, monobasic-specific aspartyl protease yapsin 2 (Mkc7p). J. Biol. Chem. 274: 24431-24437. Kostova, Z., Rancour, D.M., Menon, A.K. and Orlean, P. (2000). Photoaffinity labelling with P3-(4-azidoanilido)uridine 5'-triphosphate identifies gpi3p as the UDP-GlcNAc-binding subunit of the enzyme that catalyses formation of GlcNAc-phosphatidylinositol, the first glycolipid intermediate in glycosylphosphatidylinositol synthesis. Biochem. J. 350: 815-822. Leidich, S.D., Drapp, D.A. and Orlean, P. (1994). A conditionally lethal yeast mutant blocked at the first step in glycosyl phosphatidylinositol anchor synthesis. J. Biol. Chem. 269: 10193-10196. Liochev, S.I. and Fridovich, I. (1994). The role of O.2- in the production of HO.: in vitro and in vivo. Free Radic. Biol. Med. 16: 29-33. Low, M.G. (1989). The glycosyl-phosphatidylinositol anchor of membrane proteins. Biochim. Biophys. Acta 988: 427-454. Low, M.G. and Saltiel, A.R. (1988). Structural and functional roles of glycosyl-phosphatidylinositol in membranes. Science 239: 268-275. Lu, C.F., Kurjan, J. and Lipke, P.N. (1994). A pathway for cell wall anchorage of Saccharomyces cerevisiae alpha-agglutinin. Mol. Cell. Biol. 14: 4825-4833. Lussier, M., White, A.M., Sheraton, J., di Paolo, T., Treadwell, J., Southard, S.B., Horenstein, C.I., Chen-Weiner, J., Ram, A.F., Kapteyn, J.C., Roemer, T.W., Vo, D.H., Bondoc, D.C., Hall, J., Zhong, W.W., Sdicu, A.M., Davies, J., Klis, F.M., Robbins, P.W. and Bussey, H. (1997). Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147: 435-450. Maeda, Y., Tomita, S., Watanabe, R., Ohishi, K. and Kinoshita, T. (1998). DPM2 regulates biosynthesis of dolichol phosphate-mannose in mammalian cells: correct subcellular localization and stabilization of DPM1, and binding of dolichol phosphate. EMBO J. 17: 4920-4929. Maeda, Y., Watanabe, R., Harris, C.L., Hong, Y., Ohishi, K., Kinoshita, K. and Kinoshita, T. (2001). PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J. 20: 250-261. Martchenko, M., Alarco, A.M., Harcus, D. and Whiteway, M. (2004). Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 15: 456-467. Mayor, S. and Riezman, H. (2004). Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 5: 110-120. McConville, M.J. and Ferguson, M.A. (1993). The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem. J. 294: 305-324. McConville, M.J. and Menon, A.K. (2000). Recent developments in the cell biology and biochemistry of glycosylphosphatidylinositol lipids. Mol. Membr. Biol. 17: 1-16. McEwen, J.G., Bedoya, V., Patino, M.M., Salazar, M.E. and Restrepo, A.E. (1987). Experimental murine paracoccidioidomycosis induced by the inhalation of conidia. J. Med. Vet. Mycol. 25: 165-175. McGwire, B.S., O’Connell, W.A., Chang, K.P. and Engman, D.M. (2002). Extracellular release of the glycosylphosphatidylinositol (GPI)-linked Leishmania surface metalloprotease, gp63, is independent of GPI phopholipolysis: implications for parasite virulence. J. Biol. Chem. 277: 8802-8809. Meyer, U., Benghezal, M., Imhof, I. and Conzelmann, A. (2000). Active site determination of Gpi8p, a caspase-related enzyme required for glycosylphosphatidylinositol anchor addition to proteins. Biochemistry 39: 3461-3471. Meyer, U., Fraering, P., Bosson, R., Imhof, I., Benghezal, M., Vionnet, C. and Conzelmann, A. (2002). The glycosylphosphatidylinositol (GPI) signal sequence of human placental alkaline phosphatase is not recognized by human Gpi8p in the context of the yeast GPI anchoring machinery. Mol. Microbiol. 46: 745-748. Morsomme, P. and Riezman, H. (2002). The Rab GTPase Ypt1p and tethering factors couple protein sorting at the ER to vesicle targeting to the Golgi apparatus. Dev. Cell 2: 307-317. Moukadiri, I., Armero, J., Abad, A., Sentandreu, R. and Zueco, J. (1997). Identification of a mannoprotein present in the inner layer of the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 179: 2154-2162. Mouyna, I., Fontaine, T., Vai, M., Monod, M., Fonzi, W.A., Diaquin, M., Popolo, L., Hartland, R.P. and Latge, J.P. (2000). Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275: 14882-14889. Muhlschlegel, F.A. and Fonzi, W.A. (1997). PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol. Cell. Biol. 17: 5960-5967. Mukherjee, P.K., Seshan, K.R., Leidich, S.D., Chandra, J., Cole, G.T. and Ghannoum, M.A. (2001). Reintroduction of the PLB1 gene into Candida albicans restores virulence in vivo. Microbiology 147: 2585-2597. Muniz, M., Morsomme, P. and Riezman, H. (2001). Protein sorting upon exit from the endoplasmic reticulum. Cell 104: 313-320. Nagamine, K., Murashima, K., Kato, T., Shimoi, H. and Ito, K. (2003). Mode of alpha-amylase production by the shochu koji mold Aspergillus kawachii. Biosci. Biotechnol. Biochem. 67: 2194-2202. Nielsen, H., Engelbrecht, J., Brunak, S. and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1-6. Noverr, M.C., Cox, G.M., Perfect, J.R. and Huffnagle, G.B. (2003). Role of PLB1 in pulmonary inflammation and cryptococcal eicosanoid production. Infect. Immun. 71: 1538-1547. Ohishi, K., Inoue, N., Maeda, Y., Takeda, J., Riezman, H. and Kinoshita, T. (2000). Gaa1p and gpi8p are components of a glycosylphosphatidylinositol (GPI) transamidase that mediates attachment of GPI to proteins. Mol. Biol. Cell 11: 1523-1533. Ohishi, K., Inoue, N. and Kinoshita, T. (2001). PIG-S and PIG-T, essential for GPI anchor attachment to proteins, form a complex with GAA1 and GPI8. EMBO J. 20: 4088-4098. Okado-Matsumoto, A. and Fridovich, I. (2001). Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J. Biol. Chem. 276: 38388-38393. Orlean, P. (1997). Biogenesis of yeast cell wall and surface components. In: The Molecular Biology of Yeast Saccharomyces (Pringle, J.R., Broch, J.R. and Jones, E.W., eds.). Vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA, pp. 229-362. Peng, T., Orsborn, K.I., Orbach, M.J. and Galgiani, J.N. (1999). Proline-rich vaccine candidate antigen of Coccidioides immitis: conservation among isolates and differential expression with spherule maturation. J. Infect. Dis. 179: 518-521. Peng, T., Shubitz, L., Simons, J., Perrill, R., Orsborn, K.I. and Galgiani, J.N. (2002). Localization within a proline-rich antigen (Ag2/PRA) of protective antigenicity against infection with Coccidioides immitis in mice. Infect. Immun. 70: 3330-3335. Richard, M., Ibata-Ombetta, S., Dromer, F., Bordon-Pallier, F., Jouault, T. and Gaillardin, C. (2002). Complete glycosylphosphatidylinositol anchors are required in Candida albicans for full morphogenesis, virulence and resistance to macrophages. Mol. Microbiol. 44: 841-853. Rodriguez-Pena, J.M., Cid, V.J., Arroyo, J. and Nombela, C. (2000). A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol. Cell. Biol. 20: 3245-3255. Ross-MacDonald, P., Coelho, P.S., Roemer, T., Agarwal, S., Kumar, A., Jansen, R., Cheung, K.H., Sheehan, A., Symoniatis, D., Umansky, L., Heidtman, M., Nelson, F.K., Iwasaki, H., Hager, K., Gerstein, M., Miller, P., Roeder, G.S. and Snyder, M. (1999). Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402: 413-418. San-Blas, G., Nino-Vega, G. and Iturriaga, T. (2002). Paracoccidioides brasiliensis and paracoccidioidomycosis: molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med. Mycol. 40: 225-242. Saporito-Irwin, S.M., Birse, C.E., Sypherd, P.S. and Fonzi, W.A. (1995). PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol. Cell. Biol. 15: 601-613. Schmiel, D.H. and Miller, V.L. (1999). Bacterial phospholipases and pathogenesis. Microbes Infect. 1: 1103-1112. Smythe, J.A., Coppel, R.L., Brown, G.V., Ramasamy, R., Kemp, D.J. and Anders, R.F. (1988). Identification of two integral membrane proteins of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 85: 5195-5199. Spreghini, E., Davis, D.A., Subaran, R., Kim, M. and Mitchell, A.P. (2003). Roles of Candida albicans Dfg5p and Dcw1p cell surface proteins in growth and hypha formation. Eukaryot. Cell 2: 746-755. Spurway, T.D., Dalley, J.A., High, S. and Bulleid, N.J. (2001). Early events in glycosylphosphatidylinositol anchor addition: substrate proteins associate with the transamidase subunit gpi8p. J. Biol. Chem. 276: 15975-15982. Stambuk, B.U. and Cardoso de Almeida, M.L. (1996). An assay for glycosylphosphatidylinositol-anchor degrading phospholipases. J. Biochem. Biophys. Methods 33: 105-115. Stevens, V.L. (1995). Biosynthesis of glycosylphosphatidylinositol membrane anchors. Biochem. J. 310: 361-370. Sturtz, L.A., Diekert, K., Jensen, L.T., Lill, R. and Culotta, V.C. (2001). A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 276: 38084-38089. Sundstrom, P. (2002). Adhesion in Candida spp. Cell. Microbiol. 4: 461-469. Takahashi, M., Inoue, N., Ohishi, K., Maeda, Y., Nakamura, N., Endo, Y., Fujita, T., Takeda, J. and Kinoshita, T. (1996). PIG-B, a membrane protein of the endoplasmic reticulum with a large lumenal domain, is involved in transferring the third mannose of the GPI anchor. EMBO J. 15: 4254-4261. Terashima, H., Hamada, K. and Kitada, K. (2003). The localization change of Ybr078w/Ecm33, a yeast GPI-associated protein, from the plasma membrane to the cell wall, affecting the cellular function. FEMS Microbiol. Lett. 218: 175-180. Thompson, J.D., Gilbson, T.J., Plewniak, F., Jeanmougin, F. and Higgins, D.G. (1997). The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876-4882. Timpe, J.M., Holm, M.M., Vanlerberg, S.L., Basrur, V. and Lafontaine, E.R. (2003). Identification of a Moraxella catarrhalis outer membrane protein exhibiting both adhesin and lipolytic activities. Infect. Immun. 71: 4341-4350. Udenfriend, S. and Kodukula, K. (1995). How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu. Rev. Biochem. 64: 563-591. Vai, M., Gatti, E., Lacana, E., Popolo, L. and Alberghina, L. (1991). Isolation and deduced amino acid sequence of the gene encoding gp115, a yeast glycophospholipid-anchored protein containing a serine-rich region. J. Biol. Chem. 266: 12242-12248. van der Vaart, J.M., Caro, L.H., Chapman, J.W., Klis, F.M. and Verrips, C.T. (1995). Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 177: 3104-3110. Varki Ajit, R.C., Esko, J., Freeze, H., Hart, G. and Marth, J. (1999). Essentials of Glycobiology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA. Vidugiriene, J., Vainauskas, S., Johnson, A.E. and Menon, A.K. (2001). Endoplasmic reticulum proteins involved in glycosylphosphatidylinositol-anchor attachment: photocrosslinking studies in a cell-free system. Eur. J. Biochem. 268: 2290-2300. Vossen, J.H., Muller, W.H., Lipke, P.N. and Klis, F.M. (1997). Restrictive glycosylphosphatidylinositol anchor synthesis in cwh6/gpi3 yeast cells causes aberrant biogenesis of cell wall proteins. J. Bacteriol. 179: 2202-2209. Watanabe, R., Ohishi, K., Maeda, Y., Nakamura, N. and Kinoshita, T. (1999). Mammalian PIG-L and its yeast homologue Gpi12p are N-acetylglucosaminylphosphatidylinositol de-N-acetylases essential in glycosylphosphatidylinositol biosynthesis. Biochem. J. 339: 185-192. Watanabe, R., Murakami, Y., Marmor, M.D., Inoue, N., Maeda, Y., Hino, J., Kangawa, K., Julius, M. and Kinoshita, T. (2000). Initial enzyme for glycosylphosphatidylinositol biosynthesis requires PIG-P and is regulated by DPM2. EMBO J. 19: 4402-4411. Weisiger, R.A. and Fridovich, I. (1973). Mitochondrial superoxide dismutase. Site of synthesis and intramitochondrial localization. J. Biol. Chem. 248: 4793-4796. Yabuki, M. and Fukui, S. (1970). Presence of binding site for alpha-amylase and of masking protein for this site on mycelial cell wall of Aspergillus oryzae. J. Bacteriol. 104: 138-144. Zhu, Y., Yang, C., Magee, D.M. and Cox, R.A. (1996). Coccidioides immitis antigen 2: analysis of gene and protein. Gene 181: 121-125. Zhu, Y., Tryon, V., Magee, D.M. and Cox, R.A. (1997). Identification of a Coccidioides immitis antigen 2 domain that expresses B-cell-reactive epitopes. Infect. Immun. 65: 3376-3380. |
|