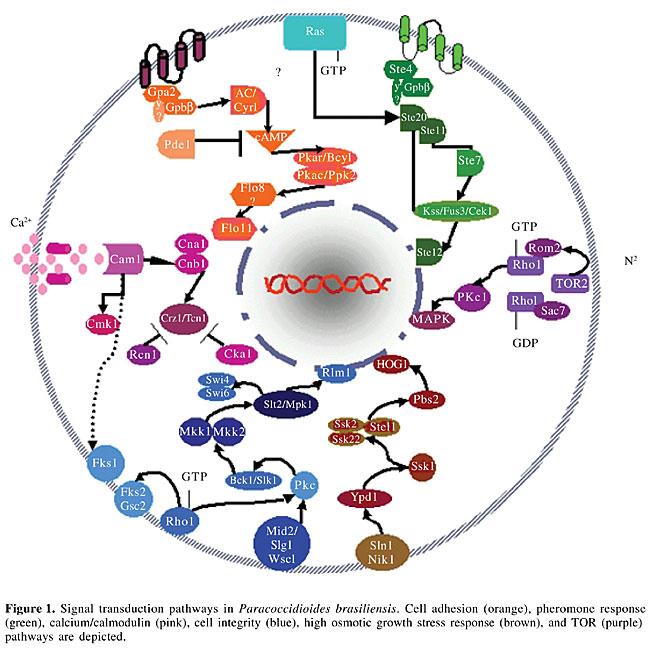
ABSTRACT. The human fungal pathogen Paracoccidioides brasiliensis is an ascomycete that displays a temperature-dependent dimorphic transition, appearing as a mycelium at 22°C and as a yeast at 37°C, this latter being the virulent form. We report on the in silico search made of the P. brasiliensis transcriptome-expressed sequence tag database for components of signaling pathways previously known to be involved in morphogenesis and virulence in other species of fungi, including Saccharomyces cerevisiae, Cryptococcus neoformans, Candida albicans, and Aspergillus fumigatus. Using this approach, it was possible to identify several protein cascades in P. brasiliensis, such as i) mitogen-activated protein kinase signaling for cell integrity, cell wall construction, pheromone/mating, and osmo-regulation, ii) the cAMP/PKA system, which regulates fungal development and virulence, iii) the Ras protein, which allows cross-talking between cascades, iv) calcium-calmodulin-calcineurin, which controls cell survival under oxidative stress, high temperature, and membrane/cell wall perturbation, and v) the target of rapamycin pathway, controlling cell growth and proliferation. The ways in which P. brasiliensis responds to the environment and modulates the expression of genes required for its survival and virulence can be inferred through comparison with other fungi for which this type of data is already available. Key words: Fungi, Paracoccidioides brasiliensis, Signaling pathways, Comparison INTRODUCTION Signal transduction describes a great number of biochemical events that transmit a signal from the cell exterior, through the cell membrane, and into the cytoplasm. This involves a number of molecules, including receptors, intermediate proteins, and messengers. The signaling pathways are commonly used by an extensive array of biological ligands to modulate various cell processes, such as growth, differentiation, and proliferation. These transduction pathways communicate information about the external environment to the inside of a cell. Signaling systems in fungi also regulate cell polarity, mating, and pheromone control, and hence they play a role in determining cell shape. Some of the known responses include changes in the cell cycle, polarized growth, and modifications to the transcriptional profile of the cell. Since the 50’s, signaling pathways have been investigated by genetic and biochemical experimentation. In a large series of experiments, eukaryotic organisms were studied for their nutritional limitations and for their reactions to various environmental stresses, such as heat, oxidative, osmotic, or ethylic shock (Engebrecht, 2003). The genomic era changed the perspective of signaling pathway studies. By using a sequence database, such as GeneBank, the screening of potentially transcribed genes in a given cell led to the rapid identification of critical genes. These methods enable researchers to assess genetic diversity or find similarities among cell types. Pathogenic organisms sense and respond to the harsh conditions imposed by the host-activating components of signaling pathways that culminate in the expression of genes responsible for the virulence and differentiation of the pathogen. Therefore, studies on signal transduction in various fungi may reveal common conserved mechanisms of signal transduction as well as the differences between these organisms. These studies could lead to drug development. We report the in silico search made of the Paracoccidioides brasiliensis transcriptome expressed sequence tag database (Felipe et al., 2003) to evaluate the presence of cellular signaling pathway elements (Figure 1) and to compare them with the cascade components in i) the non-pathogenic fungus Saccharomyces cerevisiae, ii) the human opportunistic non-dimorphic fungus Cryptococcus neoformans, iii) the human opportunistic dimorphic fungus Candida albicans, and iv) the human pathogenic fungus Aspergillus fumigatus.
MATERIAL AND METHODS The identification of putative genes involved in the cellular signaling pathways was performed by the “search by key word” service provided by the bioinformatics group of the PbGenome project (Felipe et al., 2003). The classification of candidates according to the signaling category families was performed by a BLASTx (Zhang, 2003) comparison of sequences against a database with all the signaling protein sequences from Genbank (Benson et al., 2004). The analyzed clusters were assembled by CAP3 software in the sequence-processing pipeline from the PbGenome project. The multiple sequence alignment of the candidates was performed using CLUSTAL W software (Aiyar, 2000). THE SIGNALING PATHWAYS IN PARACOCCIDIOIDES BRASILIENSIS The mitogen-activated protein kinase cascade The mitogen-activated protein kinase (MAPK) pathway is a major mechanism for controlling transcription in eukaryotes (Seger and Krebs, 1995). MAPK was originally discovered as an insulin-activated protein-serine kinase, though biochemical studies, reinforced by genetic analysis, indicate a pheromone response in budding yeast. Fungal cells contain five MAPK cascades that orchestrate responses to different physiological stimuli. One cascade operates in cells undergoing meiosis and regulates spore formation. The other four cascades operate in vegetative, mitotically active cells. Among these cascades, two control developmental events - mating and filamentation. The pheromone response pathway, the cell integrity pathway, and the high osmolarity glycerol (HOG) pathway, can all activate their MAPKs within minutes after initial stimuli (Lengeler et al., 2000). The pheromone response Pheromones are ubiquitous in fungal mating systems, and even their general structure is well conserved. Although the pheromone response pathway, and that regulating filamentous growth, share a common MAPK module, their upstream regulators appear to be specific to each of them. Haploid fungal cells respond to pheromones by a MAPK cascade involved in mating. Normal S. cerevisiae cells undergo a vegetative life cycle, but after binding of the appropriate mating pheromone they activate a different developmental pathway that leads to the production of mating filaments. This intercellular communication between the two mating types of cells activates a signal transduction pathway that stimulates the various physiological changes required by the process, such as induction of cell surface agglutinins, cell division arrest at G1, morphogenesis to form a conjugation tube, and cell fusion. The components of this cascade include a G-protein-coupled receptor, several protein kinases, and a pheromone-responsive transcription factor. The molecular mechanisms that transduce the pheromone signal are remarkably similar to the mechanisms of hormone signaling used in multicellular organisms (Konopka and Fields, 1992). The yeast protein kinases encoded by STE11, STE7, and FUS3 constitute the kinase cascade in which Ste11p phosphorylates and activates Ste7p, which in turn phosphorylates the MAPK Fus3p (Neiman and Herskowitz, 1994). In this model of signaling, specificity is defined by at least four ramifications of the same signaling pathway. Briefly, Ste12p and Mcm1p activate the transcription factor of the genes that respond to pheromones in haploid cells, whereas in diploid cells, Ste12p and Tec1p activate the genes responsible for filamentation (Lengeler et al., 2000). In C. neoformans, the STE12 homologue gene, though it has conserved roles in morphogenesis, exists only in mating type (MAT)a cells, and it is directly involved in virulence (Chang et al., 2000). Recently, three genes encoding the MF pheromone were identified in the mating-type locus, and they were found to be transcriptionally induced by limiting nutrients and to co-culture with MATa cells. Overexpression of MFa pheromone enhances haploid budding and the MFa pheromone is not essential for virulence, but it contributes to overall virulence (Wang et al, 2000). The mating process in C. albicans was cytologically described for the first time by Bennett et al. (2003). The cascade consists of Cst20p kinases, which phosphorylate Hst7p and Cek1p, which are homologous to Fus3p and Kss1p (Lockhart et al., 2002). The finding of homologue genes suggests the existence of a sexual cycle in A. fumigatus. Using the available incomplete genome database, Poggeler and Kuck (2002) deduced that the gene products of the receptor are putative proteins of seven transmembrane domains, which display a high-level amino acid identity with the a-factor receptor Ste3p and the a-receptor Ste2p of S. cerevisiae. The complete pathway from the Skh1p protein (Ste4p), which includes the downstream Ste20p and its targets in tandem Ste11p and Ste7p, was identified in P. brasiliensis. The downstream components, such as Kss1p (also called Fus3p/Cek1p), which phosphorylates Ste12p, were also identified in this transcriptome. The proteins Ste20p, Ste11p, Ste7p, and Kss1p appear to be constitutively expressed in both mycelial and yeast forms. On the other hand, genes MOT2 and MOT3, which encode nuclear Zn-finger proteins that attenuate the mating pheromone, were not found in the P. brasiliensis transcriptome. The function assigned to the whole pathway is to positively regulate growth and cell proliferation (Table 1). 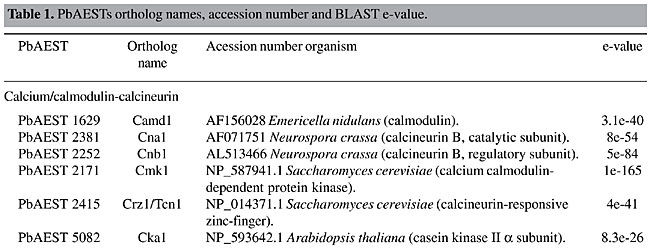  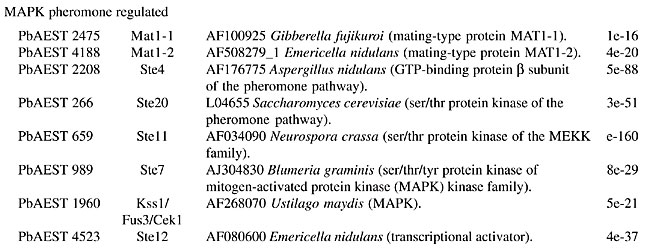 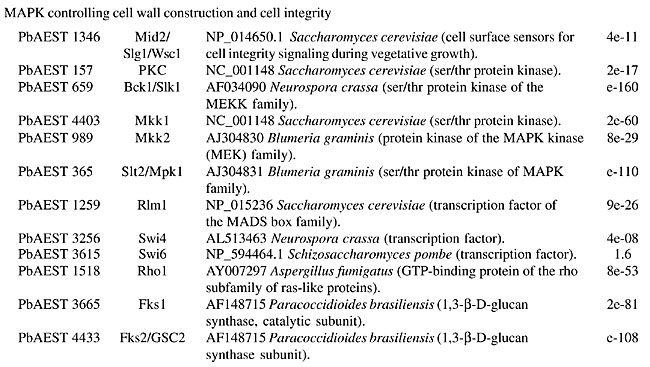  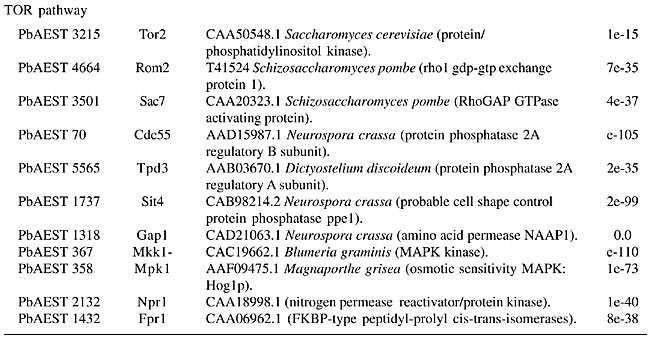 Four mating-type proteins were found through the transcriptome analyses of P. brasiliensis: a homologue to the MAT-1 protein of A. nidulans, with an E-value of 1e-46, and three PbAESTs, which possibly encode a MAT-2 (Table 1). Despite the presence of the genes involved in the cascade responsive to pheromones, a conclusive observation of the sexual cycle of P. brasiliensis is still pending; however, we present evidence that suggests the existence of mating in this organism. Maintenance of cell integrity In the budding yeast, S. cerevisiae, the MAPK cascade responsible for cell integrity, mediates cell cycle-regulation, cell wall synthesis and responds to various signals, including temperature, changes in external osmolarity, and mating pheromone. Signaling proteins found in P. brasiliensis that compose this pathway include GTP binding protein Rho1p, protein kinase C homologue, Pkc1p, MEKK Bck1p (also known as Slk1p), a redundant pair of MEKs, Mkk1p and Mkk2p, MAPK Slt2p (also called Mpk1p), and transcription factor targets Rlm1p and SBF complex (Table 1); the latter is composed of the polypeptides Swi4p and Swi6p (Gustin et al., 1998). There are probably many branches into and out of this pathway. Rho1p is a small GTP-binding protein of the Rho subfamily of Ras-related proteins that is required for cell growth. In C. neoformans, a homologue of S. cerevisiae Mpk1/Slt2 MAPK was identified by Kraus and Heitman (2003). Their work characterized the role of Mpk1p in the maintenance of cell integrity in response to elevated growth temperature and to cell-wall-synthesis inhibitors. The protein encoded by C. neoformans MPK1 is required for growth at 37°C in vitro; this growth defect is suppressed by osmotic stabilization. The cell wall is a fungus-specific dynamic structure essential to almost every aspect of the biology and pathogenicity of C. albicans. Its structure confers physical protection and shape to fungal cells, and as the most external part of the fungus, it mediates the interaction with the host, including adhesion to host tissues and modulation of the host anti-Candida immune response. Therefore, the search for potential cell wall-related targets can be envisaged as key to understanding fungal pathobiology. Moreno et al. (2003) characterized (in silico) a C. albicans gene encoding a putative transcriptional factor required for cell wall integrity. This gene codes for a Zn(II) Cys(6) transcriptional factor involved in cell wall architecture. Cell wall integrity in A. fumigatus is less well defined, and its role in protecting fungal cells is not clearly understood. In the model yeast S. cerevisiae, the cell integrity Mpk1/Slt2 MAPK and calcineurin pathways monitor the state of the cell wall and promote remodeling under stress conditions. The Rho1p of P. brasiliensis bears 86.9% identity with the S. cerevisiae counterpart. Rho1p binds to and is required for the activity of Pkc1p in vivo, which in turn regulates the MAPK pathway. In addition, Rho1p may participate in the synthesis of a-1,3 glucan, the main structural component of the yeast cell wall in P. brasiliensis. The synthesis in S. cerevisae occurs at the cell surface by action of two differentially expressed glucan synthases, Fks1p and Fks2p. For Fks1p to be active, Rho1p must be in its GTP-bound form (Gustin et al., 1998). Osmoregulation by the HOG pathway To maintain cellular homeostasis, the fungal cells need to have a way to avoid dehydration in conditions of solute excess in the extracellular environment. One strategy is the accumulation of glycerol inside the cell, a compatible substance that avoids cellular water outflow in high osmolarity conditions (Hohmann, 2002). One system of glycerol production is the HOG pathway. It uses some proteins that are sensitive to differences in the concentration of solutes in the environment and transmit signals to the cell to promote osmoadaptation. The initialization of glycerol production by HOG can be triggered by two membrane osmosensors that detect osmotic alterations and transduce signals to a series of proteins that will activate MAPK HOG1 (O’Rourke and Herskowitz, 2004). Sho1p and Sln1p, two osmosensors situated in the membrane, can detect extracellular hyperosmolarity. They transfer signals from the environment through a cascade that will activate Pbs2p (MAPKK), which phosphorylates Hog1p (Marles et al., 2004). Saccharomyces cerevisiae Sln1p is a histidine-kinase protein that contains 1,220 amino acid residues, with homologues in some fungi, such as C. albicans and P. brasiliensis. Sln1p is essential for osmoadaptation in the budding yeast S. cerevisiae, in which it acts as a negative regulator of the HOG pathway; its deletion in this organism is lethal (Posas et al., 1996). In C. albicans, Sln1p is partially required for osmoadaptation, and sln1 mutants have defective morphogenesis and are less virulent (Nagahashi et al., 1998). At a constant osmotic pressure, Sln1p inhibits the expression of Ssk1p, which is required in one branch of this pathway. Under high extracellular osmolarity, Sln1p is inhibited and Ssk1p is activated, transmitting signals to the other MAPKKs, such as Ssk2p (O’Rourke and Herskowitz, 2004). The second osmosensor is Sho1p, a protein with 367 amino acid residues, found mainly in polarized growth regions of S. cerevisiae cells. Under conditions of high osmolarity, Sho1p transfers the signal to Ste20p, a PKA that activates MAPKKK Ste11p, which is required for activation of Pbs2p. Glycerol production is triggered when the Hog1p is phosphorylated by Pbs2p and translocated to the nucleus, where it promotes the activation of several genes involved in the production of glycerol, which accumulates inside the cell (O’Rourke and Herskowitz, 2004). The in silico analysis of PbAESTs describes the components of the HOG pathway in P. brasiliensis (Table 1). The comparison of the Pb HOG proteins with their homologues in C. albicans and S. cerevisiae, as well as the multiple sequence alignment in Figure 2, which shows the high conservation of the MAPK domain and the kinase-associated domain 1 of Hog protein, all suggest that P. brasiliensis is able to use this same strategy to avoid cell dehydration.
cAMP/PKA pathway Cyclic AMP (cAMP) is the regulatory component of a well-characterized signaling pathway implicated in a variety of cellular process among fungal species. Various components of the cAMP signaling pathway were identified through transcriptome analysis of P. brasiliensis, including adenylate cyclase Cyr1p, two cAMP-dependent protein kinase catalytic subunits, Tpk1p and Tpk2p, cAMP-dependent protein kinase regulatory subunit Bcy1p, and a low affinity phosphodiesterase, Pde1p (Table 1). cAMP/protein kinase A (PKA) pathway elements have been dissected in various fungi. In C. neoformans, the cAMP signaling cascade is necessary for production of the capsule, mating filaments and melanin, all of them considered virulence factors for this organism. Mutants of the CAC1 gene for adenylate cyclase in C. neoformans are defective in capsule production and melanin synthesis; they fail to produce mating filaments and were found to be avirulent in a mouse model of cryptococcosis (Pukkila-Worley and Alspaugh, 2004). In C. albicans, adenylate cyclase - CaCDC35, PKA catalytic subunits (Tpk1p and Tpk2p) and Efg1p (hypha-specific transcriptional activator), all of them elements of the cAMP cascade, are involved in the regulation of vegetative growth and are essential for the serum-induced switch of yeast to hyphae, which has to do with the virulence of this fungus (Rocha et al. 2001; Cloutier et al., 2003). Mutants of CaCDC35 are unable to switch from yeast to hyphae, and they are unable to establish vaginal infection in a mouse model of candidiasis; they are also avirulent in systemic disease (Rocha et al., 2001). In the human pathogen A. fumigatus, elements of the cAMP pathway, such as ACYA (adenylate cyclase), PKAC and PKAR (PKA catalytic and regulatory subunits, respectively) have recently been shown to regulate the virulence gene pksP (Liebmann et al., 2003). The DacyA mutants of A. fumigatus have severely impaired sporulation and growth (Liebmann et al., 2003) and are more susceptible to being killing by human monocyte-derived macrophages. The yeast prototype, S. cerevisiae, in which the cAMP signaling pathway elements are best known. This cascade is activated by nitrogen starvation, and it transduces the signals that control the phenotypic switch in haploid cells that promote invasive growth. In diploid cells, these signals transform budding yeast into elongated forms that constitute a chain known as a pseudohypha. In addition, cAMP controls cell growth, metabolism, stress resistance, and agar adherence (Lengeler et al., 2000). The regulation of adenylate cyclase activity differs among the species. In S. cerevisiae, Ras proteins and the Ga protein GPA2 activate adenylyl cyclase in response to nutrient conditions and intracellular acidification (Colombo et al., 1998); however, in Schizosaccharomyces pombe, adenylate cyclase is regulated by Gpa2p, and Ras1p has no effect (Fukui et al., 1986). In C. neoformans and A. fumigatus, GPA1 and GPAB were identified; both of their Ga protein homologues are involved in the activation of this pathway (Madhani et al., 1999; Liebmann et al., 2003). The G-protein-coupled-receptor (Gpcr), the upstream component responsible for sensing the nitrogen limitations in S. cerevisiae (Pan et al., 2000), was also identified in P. brasiliensis, although it is still not described in C. neoformans, C. albicans and A. fumigatus. In S. cerevisiae, Cyr1p (adenylate cyclase) is an essential gene responsible for the progression of cells through the G1 phase (Mosch et al., 1999), while in C. albicans, A. fumigatus and C. neoformans, it is not an essential gene. Despite the high degree of conservation of the signaling cascade elements among fungi, their specific functions differ greatly. The multiple sequence alignment in Figure 2 shows that P. brasiliensis has two different catalytic PKA subunits, defined as Tpk1p or TPK1 (PBAEST1562) and Tpk2p or TPK2 (PBAEST3488), with a very conserved serine/threonine protein kinase catalytic domain. The enzymatic activity of these protein kinases is controlled by the phosphorylation of specific residues in the activation segment of the catalytic domain. The differences in the roles of adenylate cyclase can be explained by the fact that the cAMP pathway in S. cerevisiae is primarily involved in mediating nutritional signals to the cell cycle machinery, whereas in pathogenic or opportunistic fungi, the main function of these components is to sense and transduce stress signals from the host environment to the morphogenetic machinery that controls virulence and pathogenicity factors, thus establishing infection and allowing survival of the pathogen. In the dimorphic human pathogen Histoplasma capsulatum, it has been shown that during the transition from budding yeast to mycelium, which is dependent on a temperature shift from 37° to 25°C, there is an increase in intracellular levels of cAMP associated with this dramatic morphological switch. In addition, exogenous cAMP and phosphodiesterase inhibitors allow the Y-M transition, even at 37°C (Sacco et al., 1981). In our biological model, P. brasiliensis, evidence that cAMP has a role in the dimorphic transition, which is closely related to virulence, was first reported in 1985 by Paris and Duran, who observed that exogenous cAMP inhibits rather than induces the process of filamentation in this organism. The identification of cAMP/PKA signaling pathway components suggests a possible mechanism for cAMP involvement in the temperature-dependent dimorphic transition from mycelium to yeast. However, the complexity of the pathogenesis regulation in P. brasiliensis requires further studies of each signaling element. The roles of Ras RAS proteins belong to the Rho family - a superfamily of GTPases. It is a small guanine nucleotide-binding protein bound to the plasma membrane and involved in the regulation of several cell functions. It hydrolyses GTP; the GTP-bound form is active and GDP is inactive (Waugh et al., 2002). In human cells, this class of proteins plays a role in transmitting growth regulation signals from membrane-localized tyrosine-kinase receptors to different pathways, and ultimately to the nucleus. Some mutations in the RAS1 gene have been associated with unchecked cell growth, resulting in cancer (Barbacid, 1987). In S. cerevisiae, the Ras proteins regulate pseudohyphal growth and act upstream of the MAPK and cAMP/PKA pathways (Mösch et al., 1999). In C. neoformans, Ras1 protein is involved in filamentation, mating and high-temperature growth. The ras1 mutants are avirulent in meningitis animal models; they have a growth defect at 37°C that arrests them as large unbudded cells with depolarized actin and loss of cytoskeletal asymmetry. They are unable to produce filaments (Waugh et al., 2002). A homologue of RAS was also identified in P. brasiliensis (Table 1) and in C. albicans, where it is not an essential gene, but it is involved in hyphal growth, since ras1/ras1 mutants have a severe defect in filamentation in response to serum (Leberer et al., 2001). In C. albicans, Ras1p is also implicated in the cross talk between the MAPK and cAMP/PKA systems for the activation of the hypha-specific transcriptional factor Efg1, which is involved in the dimorphic transition from yeast to hyphae. The RAS gene should be further investigated in P. brasiliensis to elucidate its function in the Y-M transition and its possible involvement in pathogenicity. Calcium-calmodulin-calcineurin dealing with stress Among the diverse activities performed by cells, adaptation to changes in the environment is crucial for their viability. Calcium signaling is important in eukaryotes in numerous cellular processes, including the alteration of gene expression in response to external stimuli. Two important mediators of calcium signals in eukaryotic cells are the Ca2+-binding protein calmodulin (Camp) and the Ca2+/calmodulin-dependent phosphatase calcineurin. Calmodulin is present in all eukaryotic cells, including P. brasiliensis (Table 1). In the cytoplasm, it has been implicated in the regulation of protein kinases, protein phosphatases, transcription factors, motor proteins, and cytoskeletal components. Calmodulin is usually genetically represented by a single copy gene that varies from 1 to 15 kb (de Carvalho et al., 2003; Kraus and Heitman, 2003). Inhibitors of the calmodulin pathway are able to impair the transition from mycelium to yeast cells in P. brasiliensis (de Carvalho et al., 2003). In S. cerevisiae studies using temperature-sensitivity mutants of Camp, it has been found that this is an essential protein for cytoskeletal actin organization, endocytosis, and nuclear division (Desrivieres et al., 2002). In C. albicans, calcium signaling via calmodulin is important for the transition from the yeast to the filamentous form. Camp, which regulates cell proliferation and mediates the secondary messenger functions of Ca2+, was also identified in several fungi, including Phycomyces blakesleeanus, Neurospora crassa and A. nidulans. Recent studies reported that Camp plays a vital role in chitin synthase activation by mediating phosphorylation of specific microsomal protein kinases during chitin formation in N. crassa (Suresh and Subramanyam, 1997). Calcineurin is a serine-threonine-specific phosphatase that is conserved among eukaryotes. It consists of a catalytic subunit (CnaAp) and a Ca2+-binding regulatory subunit (CnaBp), and the association of the two subunits is essential for activity (Watanabe et al., 1996). Activation of calcineurin occurs when Ca2+/calmodulin binds to the C-terminal region of CnaAp, resulting in a conformational change that releases the active site from an auto-inhibitory domain. In C. neoformans, it is required for mating and haploid fruiting, for growth at elevated temperature and for virulence. Calcineurin participates in the morphogenesis of S. pombe by altering septal positioning and aberrant spindle body organization (Yoshida et al., 1994; Odom et al., 1997). Calcineurin was identified in P. brasiliensis transcriptome analysis and aligns well with its known homologues (Figure 2), which is also true for the catalytic serine/threonine domain specific to 2A phosphatases. In S. cerevisiae, the expression of the truncated CnaAp resulted in cell elongation with a unipolar budding pattern (Mendoza et al., 1996). In C. neoformans, calcineurin plays a role in hyphal elongation, observed in mating and in the survival of the heterokaryon produced by cell fusion. It is also required for hyphal elongation in diploid strains and during asexual haploid budding of MATa cells in response to nitrogen limitation (Cruz et al., 2001). Activated calcineurin dephosphorylates and activates the transcriptional factor Crz1p/Tcn1p, which enters the nucleus and, in turn, activates a set of responsive genes by binding to calcineurin-dependent responsive elements. It has been well documented that Crz1p/Tcn1p mediates most of the transcriptional responses driven by the activation of calcineurin under stress conditions (Viladevall et al., 2004). The genes that are controlled by Cna1p/Crz1p encode products that promote cell survival under stress. Crz1p is the best-characterized substrate of calcineurin in yeast, although recent studies on the identification of new substrates promise to provide novel insights into additional functions of calcineurin in vivo (Cyert, 2003). Most Tcn1p-dependent genes can be differentially induced based on mechanisms of sensitivity to Ca2+ signals and other regulatory inputs (Matheos et al., 1997). Calcium-mediated signaling mechanisms are used by virtually every eukaryotic cell to regulate a wide variety of cellular processes, including the maintenance of cell integrity under various types of stress. Paracoccidioides brasiliensis may use this machinery to protect itself against the harsh environment of the host. TOR signaling pathway TOR (target of rapamycin) is a phosphatidylinositol kinase-related protein kinase that controls cellular functions necessary for cell growth and proliferation in response to nutrients (Helliwell et al., 1998). In a recent study, it was reported that TOR acts within a growth regulatory network mediated by nutrient availability. This network affects all aspects of gene expression, including transcription, translation, and protein stability (Powers et al., 2004). In addition, the TOR pathway regulates the developmental program of pseudohyphal differentiation in concert with highly conserved MAPK and PKA signaling cascades (Rohde and Cardenas, 2004). In S. cerevisiae and S. pombe, there are two TOR proteins, Tor1p and Tor2p; however, other organisms, such as C. albicans and C. neoformans, have only one TOR homologue (Cruz et al., 1999). The function of Tor1p is conserved throughout evolution; it is involved in translation initiation, transcription, ribosome biogenesis and tRNA synthesis (Schmelzle et al., 2004). In addition, Tor2p has two functions: one that is redundant to that of Tor1p and another that is unique, controlling the organization of the actin cytoskeleton during the cell cycle. Tor2p is not sensitive to rapamycin, being inhibited only by nitrogen starvation (Rohde and Cardenas, 2004). Tor2p activates Rho1p GTPase, exchanging the GDP of the inactive form for GTP. This exchange is achieved by Rom2p protein, a guanidine nucleotide exchange factor, whereas Sac7p is a GTPase-activating protein. Rho1p phosphorylates Pkc1p, which activates the MAPK cascade Bck1p, Mkk1/2p and Mpk1p (Hay and Sonenberg, 2004). In the course of P. brasiliensis transcriptome analysis, TOR2, ROM2, RHO1, SAC7, PKC1, BCK1, MKK1, MPK1, and FPR1 homologues were identified (Table 1). Although Tor1p was not identified, some components of its signaling pathway, such as Cdc55p, Tpd3p and Sit4p are present in P. brasiliensis. Therefore, Tor1p may be present in the P. brasiliensis genome, unless Tor2p fulfils both functions in P. brasiliensis. Multiple sequence analysis of Tor2p revealed a kinase domain (Figure 2). Tor protein has a C-terminal region homology with the catalytic domain of phosphatidylinositol 3-kinase and phosphatidylinositol 4-kinase. This domain is characteristic of the TOR family and is essential to its function (Crespo and Hall, 2002). CONCLUSIONS Transcriptome analyses are a powerful tool to investigate organisms such as P. brasiliensis, in which the direct genetic analysis has many drawbacks and for which there is no efficient method of direct genetic analysis. By this comparative approach, it was possible to identify several components of signaling pathways in P. brasiliensis known to regulate cell events, such as morphogenesis and virulence. These components were found to be similar to those of other fungi. We suggest that P. brasiliensis has co-opted a conserved signaling pathway to regulate phenotypes required for virulence, as have the other pathogenic microorganisms. However, there are still regulatory connections that need to be understood, in particular the broad transcriptional effects caused by the loss of the aforementioned cascades. The identification of pathogen-specific signaling events will help us understand P. brasiliensis pathogenesis, which could lead to the development of new antifungals. ACKNOWLEDGMENTS Research supported by MCT/CNPq, CNPq, Capes, FUB, UFG. We thank Dr. Renata C. Pascon for critical reading the manuscript and Hugo Costa Paes for English revision. REFERENCES Aiyar, A. (2000). The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132: 221-241. Barbacid, M. (1987). ras genes. Annu. Rev. Biochem. 56: 779-827. Bennett, R.J., Uhl, M.A., Miller, M.G. and Johnson, A.D. (2003). Identification and characterization of a Candida albicans mating pheromone. Mol. Cell Biol. 23: 8189-8201. Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J. and Wheeler, D.L. (2004). GenBank: update. Nucleic Acids Res. 32: D23-D26. Chang, Y.C., Wickes, B.L., Miller, G.F., Penoyer, L.A. and Kwon-Chung, K.J. (2000). Cryptococcus neoformans STE12alpha regulates virulence but is not essential for mating. J. Exp. Med. 191: 871-882. Cloutier, M., Castilla, R., Bolduc, N., Zelada, A., Martineau, P., Bouillon, M., Magee, B.B., Passeron, S., Giasson, L. and Cantore, M.L. (2003). The two isoforms of the cAMP-dependent protein kinase catalytic subunit are involved in the control of dimorphism in the human fungal pathogen Candida albicans. Fungal Genet. Biol. 38: 133-141. Colombo, S., Ma, P., Cauwenberg, L., Winderickx, J., Crauwels, M., Teunissen, A., Nauwelaers, D., de Winde, J.H., Gorwa, M.F., Colavizza, D. and Thevelein, J.M. (1998). Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 17: 3326-3341. Crespo, J.L. and Hall, M.N. (2002). Elucidating TOR signalling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66: 579-591. Cruz, M.C., Cavallo, L.M., Gorlach, J.M., Cox, G., Perfect, J.R., Cardenas, M.E. and Heitman, J. (1999). Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologues in Cryptococcus neoformans. Mol. Cell Biol. 19: 4101-4112. Cruz, M.C., Fox, D.S. and Heitman, J. (2001). Calcineurin is required for hyphal elongation during mating and haploid fruiting in Cryptococcus neoformans. EMBO J. 20: 1020-1032. Cyert, M.S. (2003). Calcineurin signalling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311: 1143-1150. de Carvalho, M.J., Amorim-Jesuino, R.S., Daher, B.S., Silva-Pereira, I., de Freitas, S.M., Soares, C.M. and Felipe, M.S. (2003). Functional and genetic characterization of calmodulin from the dimorphic and pathogenic fungus Paracoccidioides brasiliensis. Fungal Genet. Biol. 39: 204-210. Desrivieres, S., Cooke, F.T., Morales-Johansson, H., Parker, P.J. and Hall, M.N. (2002). Calmodulin controls organization of the actin cytoskeleton via regulation of phosphatidylinositol (4,5)-bisphosphate synthesis in Saccharomyces cerevisiae. Biochem. J. 366: 945-951. Engebrecht, J. (2003). Cell signalling in yeast sporulation. Biochem. Biophys. Res. Commun. 306: 325-328. Felipe, M.S., Andrade, R.V., Petrofeza, S.S., Maranhao, A.Q., Torres, F.A., Albuquerque, P., Arraes, F.B., Arruda, M., Azevedo, M.O., Baptista, A.J., Bataus, L.A., Borges, C.L., Campos, E.G., Cruz, M.R., Daher, B.S., Dantas, A., Ferreira, M.A., Ghil, G.V., Jesuino, R.S., Kyaw, C.M., Leitao, L., Martins, C.R., Moraes, L.M., Neves, E.O., Nicola, A.M., Alves, E.S., Parente, J.A., Pereira, M., Pocas-Fonseca, M.J., Resende, R., Ribeiro, B.M., Saldanha, R.R., Santos, S.C., Silva-Pereira, I., Silva, M.A., Silveira, E., Simoes, I.C., Soares, R.B., Souza, D.P., De-Souza, M.T., Andrade, E.V., Xavier, M.A., Veiga, H.P., Venancio, E.J., Carvalho, M.J., Oliveira, A.G., Inoue, M.K., Almeida, N.F., Walter, M.E., Soares, C.M. and Brigido, M.M. (2003). Transcriptome characterization of the dimorphic and pathogenic fungus Paracoccidioides brasiliensis by EST analysis. Yeast 20: 263-271. Fukui, Y., Kozasa, T., Kaziro, Y., Takeda, T. and Yamamoto, M. (1986). Role of a ras homologue in the life cycle of Schizosaccharomyces pombe. Cell 44: 329-336. Gustin, M.C., Albertyn, J., Alexander, M. and Davenport, K. (1998). MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62: 1264-1300. Hay, N. and Sonenberg, N. (2004). Upstream and downstream of mTOR. Genes Dev. 18: 1926-1945. Helliwell, S.B., Howald, I., Barbet, N. and Hall, M.N. (1998). TOR2 is part of two related signalling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics 148: 99-112. Hohmann, S. (2002). Osmotic adaptation in yeast-control of the yeast osmolyte system. Int. Rev. Cytol. 215: 149-187. Konopka, J.B. and Fields, S. (1992). The pheromone signal pathway in Saccharomyces cerevisiae. Antonie van Leeuwenhoek 62: 95-108. Kraus, P.R. and Heitman, J. (2003). Coping with stress: calmodulin and calcineurin in model and pathogenic fungi. Biochem. Biophys. Res. Commun. 311: 1151-1157. Leberer, E., Harcus, D., Dignard, D., Johnson, L., Ushinsky, S., Thomas, D.Y. and Schroppel, K. (2001). Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42: 673-687. Lengeler, K.B., Davidson, R.C., D’Souza, C., Harashima, T., Shen, W.C., Wang, P., Pan, X., Waugh, M. and Heitman, J. (2000). Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64: 746-785. Liebmann, B., Gattung, S., Jahn, B. and Brakhage, A.A. (2003). cAMP signalling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol. Genet. Genomics 269: 420-435. Lockhart, S.R., Pujol, C., Daniels, K.J., Miller, M.G., Johnson, A.D., Pfaller, M.A. and Soll, D.R. (2002). In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162: 737-745. Madhani, H.D., Galitski, T., Lander, E.S. and Fink, G.R. (1999). Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signalling mutants. Proc. Natl. Acad. Sci. USA 96: 12530-12535. Marles, J.A., Dahesh, S., Haynes, J., Andrews, B.J. and Davidson, A.R. (2004). Protein-protein interaction affinity plays a crucial role in controlling the sho1p-mediated signal transduction pathway in yeast. Mol. Cell 14: 813-823. Matheos, D.P., Kingsbury, T.J., Ahsan, U.S. and Cunningham, K.W. (1997). Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 11: 3445-3458. Mendoza, I., Quintero, F.J., Bressan, R.A., Hasegawa, P.M. and Pardo, J.M. (1996). Activated calcineurin confers high tolerance to ion stress and alters the budding pattern and cell morphology of yeast cells. J. Biol. Chem. 271: 3061-3067. Moreno, I., Pedreno, Y., Maicas, S., Sentandreu, R., Herrero, E. and Valentin, E. (2003). Characterization of a Candida albicans gene encoding a putative transcriptional factor required for cell wall integrity. FEMS Microbiol. Lett. 226: 159-167. Mösch, H.U., Kubler, E., Krappmann, S., Fink, G.R. and Braus, G.H. (1999). Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10: 1325-1335. Nagahashi, S., Mio, T., Ono, N., Yamada-Okabe, T., Arisawa, M., Bussey, H. and Yamada-Okabe, H. (1998). Isolation of CaSLN1 and CaNIK1, the genes for osmosensing histidine kinase homologues, from the pathogenic fungus Candida albicans. Microbiology 144: 425-432. Neiman, A.M. and Herskowitz, I. (1994). Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue STE7 by STE11. Proc. Natl. Acad. Sci. USA 91: 3398-3402. Odom, A., Muir, S., Lim, E., Toffaletti, D.L., Perfect, J. and Heitman, J. (1997). Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16: 2576-2589. O’Rourke, S.M. and Herskowitz, I. (2004). Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell. 15: 532-542. Pan, X., Harashima, T. and Heitman, J. (2000). Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 3: 567-572. Paris, S. and Duran, S. (1985). Cyclic adenosine 3',5' monophosphate (cAMP) and dimorphism in the pathogenic fungus Paracoccidioides brasiliensis. Mycopathologia 92: 115-120. Poggeler, S. and Kuck, U. (2001). Identification of transcriptionally expressed pheromone receptor genes in filamentous ascomycetes. Gene 280: 9-17. Posas, F., Wurgler-Murphy, S.M., Maeda, T., Witten, E.A., Thai, T.C. and Saito, H. (1996). Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86: 865-875. Powers, T., Dilova, I., Chen, C.Y. and Wedaman, K. (2004). Yeast TOR signalling: a mechanism for metabolic regulation. Curr. Top. Microbiol. Immunol. 279: 39-51. Pukkila-Worley, R. and Alspaugh, J.A. (2004). Cyclic AMP signalling in Cryptococcus neoformans. FEMS Yeast Res. 4: 361-367. Rocha, C.R., Schroppel, K., Harcus, D., Marcil, A., Dignard, D., Taylor, B.N., Thomas, D.Y.,Whiteway, M. and Leberer, E. (2001). Signalling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12: 3631-3643. Rohde, J.R. and Cardenas, M.E. (2004). Nutrient signalling through TOR kinases controls gene expression and cellular differentiation in fungi. Curr. Top. Microbiol. Immunol. 279: 53-72. Sacco, M., Maresca, B., Kumar, B.V., Kobayashi, G.S. and Medoff, G. (1981). Temperature- and cyclic nucleotide-induced phase transitions of Histoplasma capsulatum. J. Bacteriol. 146: 117-120. Schmelzle, T., Beck, T., Martin, D.E. and Hall, M.N. (2004). Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24: 338-351. Seger, R. and Krebs, E.G. (1995). The MAPK signalling cascade. FASEB J. 9: 726-735. Suresh, K. and Subramanyam, C. (1997). A putative role for calmodulin in the activation of Neurospora crassa chitin synthase. FEMS Microbiol. Lett. 150: 95-100. Viladevall, L., Serrano, R., Ruiz, A., Domenech, G., Giraldo, J., Barcelo, A. and Arino, J. (2004). Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J. Biol. Chem. 15; 279 (42): 43614-43624. Epub 2004 Aug. 6. Wang, P., Perfect, J.R. and Heitman, J. (2000). The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. 20: 352-362. Watanabe, Y., Perrino, B.A. and Soderling, T.R. (1996). Activation of calcineurin A subunit phosphatase activity by its calcium-binding B subunit. Biochemistry 35: 562-566. Waugh, M.S., Nichols, C.B., DeCesare, C.M., Cox, G.M., Heitman, J. and Alspaugh, J.A. (2002). Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148: 191-201. Yoshida, T., Toda, T. and Yanagida, M. (1994). A calcineurin-like gene ppb1+ in fission yeast: mutant defects in cytokinesis, cell polarity, mating and spindle pole body positioning. J. Cell Sci. 107: 1725-1735. Zhang, H. (2003). Alignment of BLAST high-scoring segment pairs based on the longest increasing subsequence algorithm. Bioinformatics 19: 1391-1396. |
|