ABSTRACT. Esterase (Est) and esterase-D (Est-D) electrophoretic patterns identified by starch gel electrophoresis of skeletal muscle protein extracts of 184 specimens of three species of peacock bass, locally known as tucunarés (Cichla monoculus, C. temensis and Cichla sp), plus four specimens of a supposed hybrid (C. monoculus vs C. temensis), collected from the Central Amazon, were examined to determine if they could aid in identifying a supposed hybrid between C. monoculus and C. temensis. Six zones of electrophoretic activity were found with these enzyme systems. The Est enzyme showed one zone of activity, formed by bands 1, 2 and 3, plus three zones of activity, presumably controlled by Est-1, 2 and 3 loci. The Est-D enzyme had two zones of activity, presumably controlled by Est-D1 and Est-D2 loci. Cichla monoculus and C. temensis shared band 2 and alleles Est-11, Est-21, Est-32, and Est-D11, and therefore these were useless for identifying hybrids between the two species. However, a probable hybrid pattern of bands 1, 2, and 3, presumably generated by a combination of pattern 12 from C. monoculus with pattern 23 from C. temensis, resulting from a possible cross between these two species, was detected. Although the Est-D2 locus cannot be considered an ideal diagnostic marker for identifying the supposed hybrid (C. monoculus vs C. temensis), as it is polymorphic, it proved to be useful for determining the origin of the hybrid, i.e., which parental species were involved in the hybridization process. Key words: Hybrid, Tucunarés, Esterases, Peacock bass INTRODUCTION Peacock bass, locally known as tucunarés, fish belonging to the order Perciformes, family Cichlidae, genus Cichla, are very valuable in socio-economic terms, being among the most common fish species caught and marketed in the Amazonian region. The genus Cichla is represented by five species: Cichla ocellaris Scheneider 1801, in the Venezuelan Amazon; Cichla monoculus Spix 1831, possibly throughout the Central Amazon, Cichla temensis Humboldt 1833, found in the Orinoco, Negro and Tapajós Rivers, Cichla orinocencis Humboldt 1833, in the Orinoco and Negro Rivers, and Cichla intermedia Machado-Allison 1971, in the Casiquiare River (upper Negro River) and mid Orinoco River (Kullander, 1983, 1986). However, in addition to these previously described five species, there may be others in the region, with the possibility of their total number reaching up to 15 (Ferreira, E.J.G., personal communication). Inter-specific hybridization in nature, is considered a very common phenomenon among several groups of fish, where specifically among the freshwater fish species, this phenomenon is related to environmental factors, decreasing gradually from the northern temperate to the southern tropical zones (Hubbs, 1955). According to Hubbs (op. cit.), the natural hybridization frequency appears to be inversely related to the number of species in determined areas, as is the case for the tropical region, with its highly diversified fauna, where hybridization between species tends to be less frequent. In this context, a low frequency of natural hybrids may be expected in the Amazon, even though one cannot discard the possibility that some hybrids may have been identified as distinct species in this region (Ribeiro, 1985). In the Amazon, the identification of natural hybrids in fish has already been made based on morphological studies of jaraquis (Semaprochilodus insignis vs Semaprochilodus taeniurus) (Ribeiro, op. cit.). And more recently, a hypothesis of natural hybridization in peacock bass (C. monoculus vs C. temensis) has been put forth, based on karyotypic (Alves and Feldberg, 1998) and mitochondrial DNA sequencing analyses (Andrade et al., 2001). The discrimination of electrophoretic protein band patterns has been highly practical for the identification of fish hybrids (Nyman, 1967, 1970; Brassington and Ferguson, 1976; Child and Solomon, 1977; Solomon and Child, 1978; Ferguson, 1980), yielding relevant findings on the species taxonomic status definition. We examined the electrophoretic esterase patterns of peacock bass (C. monoculus, C. temensis and Cichla sp) as an auxiliary tool for identifying a supposed hybrid between C. monoculus and C. temensis. MATERIAL AND METHODS Collection of material One hundred and eighty-four specimens of peacock bass, identified as three species of Cichla, were collected from five areas in the Central Amazon (Figure 1). Of the 108 specimens of Cichla monoculus, 33 were from Lago Catalão, at the confluence of the Rio Negro with the Rio Solimões (8 caught on 09/18/1996 and 25 on 09/27/2002); 18 were from Lago Tefé-Rio Tefé caught on 11/22/1996; 27 from Lago do Acará-Rio Japurá caught on 11/25/1996, and 30 from Lago de Balbina-Rio Uatumã caught on 12/04/1997. Of the 20 specimens of Cichla temensis, two were caught downstream from the Balbina hydroelectric power plant dam on 08/25/1997, and 18 were from Lago Marajá-Rio Negro (10 caught on 04/10/2003 and eight on 04/24/2003). Of the 56 specimens of Cichla sp at Lago de Balbina-Rio Uatumã, 25 were caught on 12/07/1997 and 31 on 09/06/2002. Additionally, four specimens, which in the course of the analyses presented a supposed hybrid electrophoretic pattern between Cichla monoculus and Cichla temensis, were included in the study, one of them being collected as Cichla temensis downstream at the Balbina hydroelectric power plant dam on 08/25/1997, and three as Cichla monoculus, from Lago Catalão on 09/27/2002.
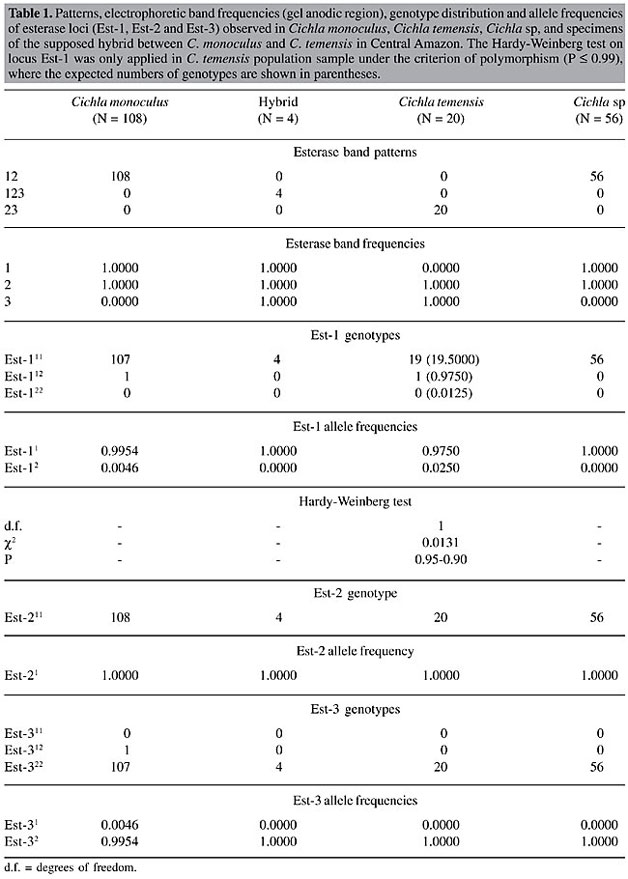
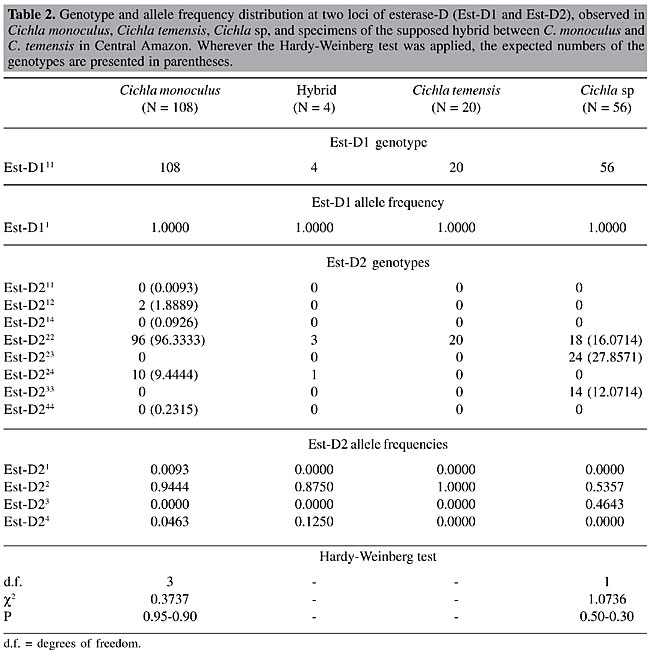
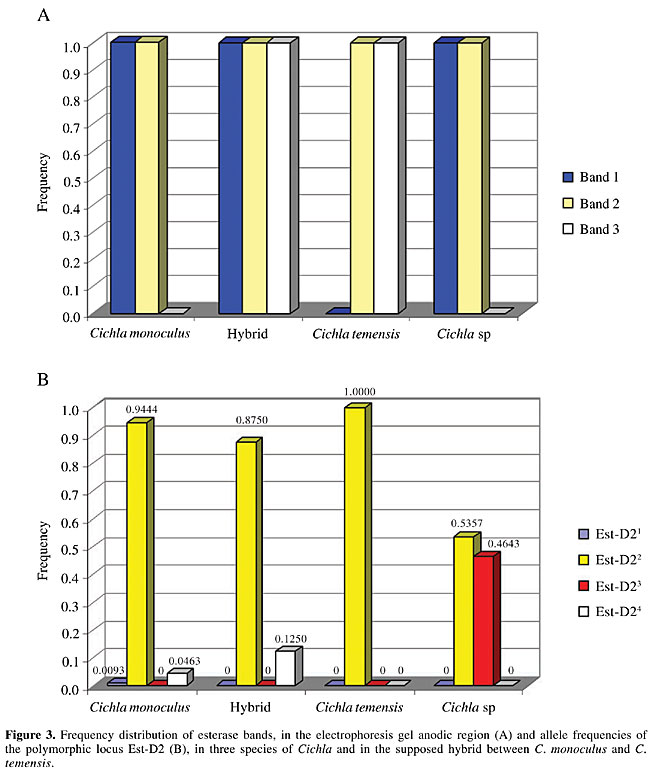
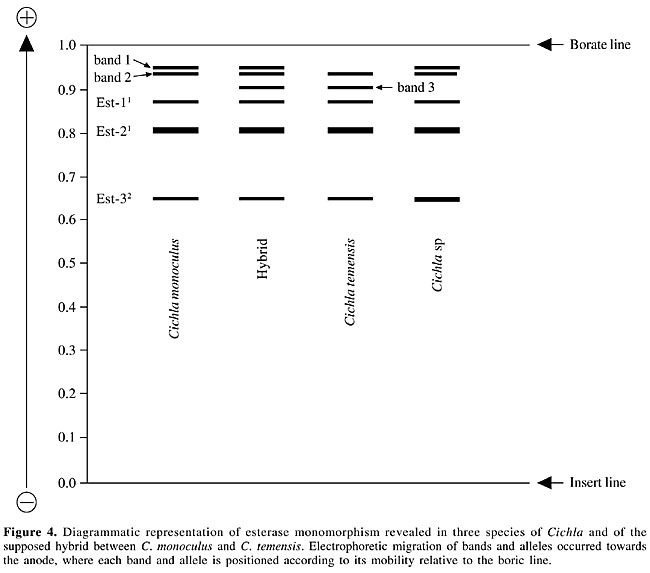
Electrophoresis Starch gel electrophoresis of the protein extracts of the skeletal muscle of the peacock bass specimens was used for discriminating the isoenzyme patterns of the enzymes esterase and esterase-D. The protein extracts were obtained by grinding equal parts of muscle and 0.10 M Tris HCl, pH 7.10, containing 1% b-mercaptoethanol. The samples were centrifuged at 3,000 rpm for 20 min just prior to the electrophoretic run in order to obtain the protein supernatants. The gel and electrode electrophoretic buffers were prepared according to the methodology described by Ridgway et al. (1970), with Sigma starch gels at 13%, submitted to a 150-V potential for 4 h. The esterase was stained according to the method of Shaw and Prasad (1970), and esterase-D was stained according to the method of Hopkinson et al. (1973). Loci, bands and alleles were identified and numerically ranked in decreasing order according to their electrophoretic migration towards the anode. Statistical analysis Chi-square (c2) tests, assuming Hardy-Weinberg equilibrium, were applied for comparing the observed and expected numbers of the esterase genotypes in the population samples of Cichla monoculus and Cichla sp. The computer program Tools for Population Genetic Analyses (TFPGA; Miller, 1997) was used for the analyses. RESULTS AND DISCUSSION The identification of hybrids by means of electrophoretic technique is based on the basic principle that an isoenzyme or protein pattern of a hybrid is formed by the sum of the bands of the patterns found for the two parental species (Nyman, 1967, 1970; Brassington and Ferguson, 1976; Child and Solomon, 1977; Solomon and Child, 1978; Ferguson, 1980). For example, if the two parental species give two fixed bands or alleles with different electrophoretic mobilities for a given protein, then a hybrid specimen will reveal both bands or alleles, usually with half the concentrations shown in the parental species (Nyman, 1970; Ferguson, 1980). In this context, the finding of monomorphic loci for different species, indicating specific fixed alleles, would greatly facilitate the identification of hybrids between Cichla monoculus and Cichla temensis. We found six zones of electrophoretic activity for the esterase and esterase-D patterns in the C. monoculus, C. temensis and Cichla sp samples. The esterase enzyme gave a single zone of activity, composed of bands 1, 2 and 3, plus three zones of activity, presumably controlled by three monomorphic loci, Est-1, 2 and 3 in all species examined (according to the 95% criterion of polymorphism, Table 1). Nevertheless under the 99% criterion of polymorphism the Est-1 locus appears to be polymorphic in C. temensis. Esterase-D revealed two zones of activity, presumably controlled by the monomorphic locus Est-D1 in all species examined and by the Est-D2 locus, which showed monomorphism in C. temensis and polymorphism in C. monoculus and Cichla sp (Table 2, Figure 2, Figure 3B). Cichla monoculus and C. temensis shared band 2 and alleles Est-11, Est-21, Est-32, and Est-D11 (Tables 1 and 2); therefore, they were useless for identifying hybrids between these two species. Yet, a probable hybrid pattern of bands 1, 2, and 3 was detected in a zone of esterase activity situated in the anodic region of the gel (Figure 4), from a specimen originally identified as C. temensis collected downstream from the Balbina Hydroelectric Power Plant Dam on 08/25/1997, and from three others originally identified as C. monoculus collected from Lago Catalão at the confluence of the Rio Negro with the Rio Solimões on 09/27/2002. Visually, these patterns, which were possibly generated by a combination of pattern 12 of C. monoculus with pattern 23 of C. temensis, as a result of intercrossing between these two species (Figure 3A, Table 1), appeared to have about half the concentrations found in the parental species.
The possibility of this being a hybrid pattern is reinforced by the fact that it was not detected in the samples of C. monoculus collected at Lago Tefé-Rio Tefé and Lago do Acará-Rio Japurá, where there is no record of C. temensis living in sympatry with C. monoculus. However, we need to account for the hybrid specimens found in Lago Catalão, since this is not considered to be an area of sympatry for these species. A plausible explanation for that would be that these species have intercrossed in other regions, such as: 1) upstream in the Rio Negro, or 2) in the vicinity of the confluence of the Rio Negro with the Rio Solimões, and then the hybrid specimens later reached Lago Catalão. A possible hybridization between Cichla sp and C. temensis could also be expected, since the esterase pattern found in Cichla sp, was the same as that in C. monoculus, i.e., pattern 12. Nevertheless, data on the allele distributions of the polymorphic locus Est-D2 of the three Cichla species (Table 2, Figure 3B) make us reject this possibility, since the electrophoretic patterns of the supposed hybrids described for this locus only revealed genotypes Est-D222 and Est-D224, with no genotype containing the common and specific allele of Cichla sp: Est-D23 (Figure 2) being detected. Only the hybrid genotype Est-D223, resulting from the combination of the fixed Est-D22 allele found in Cichla temensis with the Est-D23 allele found in Cichla sp would be indicative of a hybrid between these species. Given that the electrophoretic typing of the alleles on locus Est-D2 of these species has been correctly performed, and based on the allele frequencies of C. monoculus and C. temensis, the genotypes Est-D212, Est-D222 and Est-D224 in a supposed F1 hybrid of those two species would be expected to appear in the following proportions: 0.0093, 0.9444 and 0.0463, respectively. However, this locus cannot be considered as an ideal diagnostic marker for the identification of the supposed hybrid, as it is polymorphic; on the other hand, it was useful for the definition of its origin, i.e., which parental species participated in the hybridization process. The Hardy-Weinberg test applied for examining the genotype distribution of locus Est-1 under the 99% criterion of polymorphism revealed a good population genetic balance in C. temensis ( If the hybrid electrophoretic pattern of esterase bands 1, 2 and 3 were found in the area of sympatry of C. monoculus and C. temensis, then the hypothesis of a natural hybrid between these two species based on citogenetic (Alves and Feldberg, 1998), and mitochondrial DNA sequencing (Andrade et al., 2001) analyses would be supported. In order to do so, a larger sampling of supposed hybrids between these two species of Cichla would need to be examined. ACKNOWLEDGMENTS Research supported by the National Institute for Research in the Amazon (INPA), through the Research Institutional Projects (PPI 3-3270 and PPI 1-3090) and by the Technological and Scientific National Council (CNPq) by means of the Post-Graduation North Program (PNOPG) - Project # 550703/01-2. The authors are indebted to Mr. J.C.P. Raposo for his help with the electrophoretic analyses. REFERENCES Alves, M.N. and Feldberg, E. (1998). Análise cariotípica no gênero Cichla e considerações evolutivas na família Cichlidae. VII Simpósio de Citogenética e Evolução Aplicada de Peixes Neotropicais, Londrina, PR, Brasil, C17. Andrade, F., Schneider, H., Farias, I., Feldberg, E. and Sampaio, I. (2001). Análise filogenética de duas espécies simpátricas de tucunaré (Cichla, Perciformes), com registro de hibridização em diferentes pontos da bacia Amazônica. Rev. Virtual Iniciação Cient. UFPA 1: 1-11 (http://www.ufpa.br/propesp/Revistaic/atualizacao1/edicoes_anteriores.htm). Accessed February 1, 2001. Brassington, R.A. and Ferguson, A. (1976). Electrophoretic identification of Roach (Rutilus rutilus L.), Rudd (Scardinius erythrophthalmus L.), Bream (Abramis brama L.) and their natural hybrids. J. Fish Biol. 9: 471-477. Child, A.R. and Solomon, D.J. (1977). Observations on morphological and biochemical features of some cyprinid hybrids. J. Fish Biol. 11: 125-131. Ferguson, A. (1980). Biochemical Systematics and Evolution. Blackie, Glasgow, London, England. Hopkinson, D.A., Mestriner, M.A., Cortner, J. and Harris, H. (1973). Esterase-D: a new human polymorphism. Ann. Hum. Genet. 37: 119-137. Hubbs, C.L. (1955). Hybridization between fish species in nature. Syst. Zool. 4: 1-20. Kullander, S.O. (1983). Taxonomic studies on the percoid freshwater fish family Cichlidae in South America. PhD thesis, University of Stockholm, Stockolm, Sweden. Kullander, S.O. (1986). Cichlid Fishes of Amazon River Drainage of Peru. Swedish Museum of Natural History, Stockholm, Sweden. Miller, M.P. (1997). Tools for population genetic analyses (TFPGA) 1.3: A Windows program for the analysis of allozyme and molecular population genetic data. Computer software distributed by the author. Copright (C) 1997 Mark P. Miller. Department of Biological Sciences, Nothern Arizona University, Box 5640, Flagstaff, AZ 86011-5640. E-mail: [email protected]. Nyman, O.L. (1967). Protein variation in Salmonidae. Rep. Inst. Freshwat. Res. Drottningholm. 47: 5-38. Nyman, O.L. (1970). Electrophoretic analysis of hybrids between salmon (Salmo salar) and trout (S. trutta). Trans. Am. Fish. Soc. 99: 229-236. Ribeiro, M.C.L.B. (1985). A natural hybrid between two tropical fishes: Semaprochilodus insignis vs Semaprochilodus taeniurus (Teleostei, Characoidei, Prochilodontidae). Rev. Bras. Zool. 2: 419-421. Ridgway, G.J., Sherburne, S.W. and Lewis, R.D. (1970). Polymorphism in the esterases of Atlantic herring. Trans. Am. Fish. Soc. 99: 147-151. Shaw, C.R. and Prasad, R. (1970). Starch gel electrophoresis of enzymes: a compilation of recipes. Biochem. Genet. 4: 297-320. Solomon, D.J. and Child, A.R. (1978). Identification of juvenile natural hybrids between Atlantic Salmon (Salmo salar L.) and trout (Salmo trutta L.). J. Fish Biol. 12: 499-501. |
|