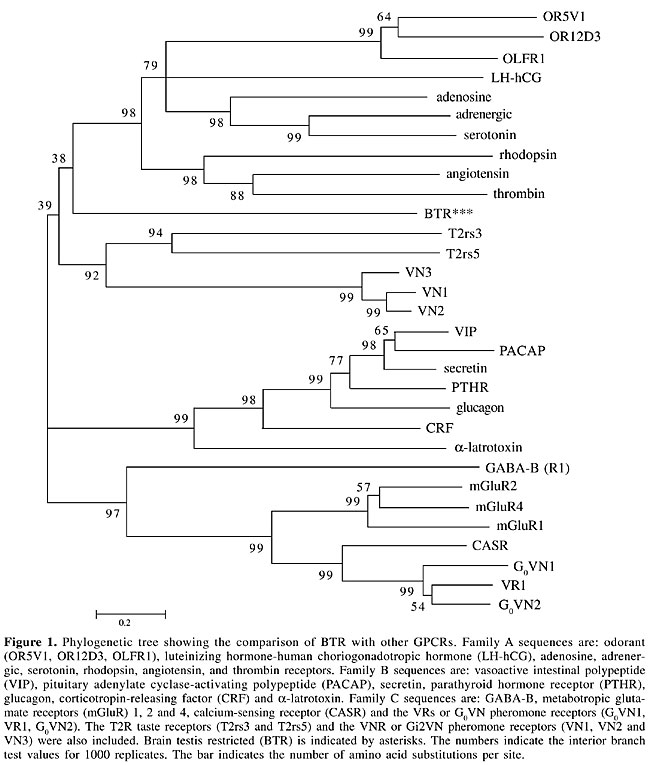
ABSTRACT. G protein-coupled receptors (GPCRs) are involved in a large variety of physiological functions. The number of known members that belong to this large family of receptors has been rapidly increasing. Now, with the availability of the human genome sequence databases, further family members are being identified. We describe the identification of a novel GPCR that shows no significant amino acid identity to any one of the known members of the GPCR superfamily. The gene expression pattern of this receptor is restricted: in normal tissues it is confined to the nervous system and testis, but we also detected gene expression in several tumor types, most notably prostate cancer, suggesting a potential role for this gene as a marker for this disease. Key words: GPCR, Brain, Testis, Prostate cancer, Orphan receptor, Bioinformatics INTRODUCTION G protein-coupled receptors (GPCRs) comprise the largest family of surface molecules involved in signal transduction. They are activated by a large variety of ligands, including hormones, growth factors, light, peptides, neurotransmitters, nucleotides and odorants, and are involved in the regulation of a range of cellular responses (Bockaert and Pin, 1999; Pierce et al., 2002). All GPCRs have a common central core domain consisting of seven transmembrane helices connected by three intracellular loops and three extracellular loops. The lengths of the N-terminal and C-terminal domains are variable in different GPCRs. The large number of GPCRs and their involvement in human diseases has made these receptors attractive drug targets. According to a recent analysis, over 30% of marketed drugs are active against this receptor super-family and yet, only a small fraction of the known GPCRs are at present drug targets (Drews, 2000). Over the last 10 years, a large number of novel GPCRs have been identified, mostly through homology cloning. The ligands remain unknown for most of these receptors (Marchese et al., 1999; Howard et al., 2001; Lee et al., 2002). Many GPCR genes do not have introns, facilitating their identification within the recently available human genome sequences (I.H.G.S.C., 2001; Venter et al., 2001). Some of the newly discovered genes are more closely related to previously known GPCRs and are likely to be activated by similar ligands. Others are more distantly related to known GPCRs and may have quite different functions. The identification of novel GPCRs, and their respective ligands, will continue to contribute to the understanding and identification of new signaling pathways involved in different aspects of human physiology. Of particular interest are receptors with recognized expression in the central nervous system, given that many psychiatric and neurodegenerative disorders are mediated by unknown mechanisms. We describe a novel putative GPCR from the human genome sequence database and characterize its expression pattern in normal human tissues as well as in tumor cell lines and tissues. The receptor was named brain testis restricted (BTR), due to its restricted expression in normal tissues. We also detected expression of the BTR gene in tumor cell lines and tissues, predominantly in prostate cancer. There is a significant correlation between BTR gene expression and prostate cancer, suggesting a potential role for BTR as a marker for detection of this type of cancer. MATERIAL AND METHODS Sequence analysis The human genome database (htgs, NCBI) was searched with consensus motifs common to members of odorant receptors (MAYDRYVAIC and KAFSTCASH) using TBLASTN. A genomic sequence containing an open reading frame (ORF) of 1044 bp was identified. The amino acid sequence was analyzed using tools in the protein fingerprint database (PRINTS) (http://www.bioinf.man.ac.uk/dbbrowser/PRINTS), PROSITE (http://www.expasy.org/prosite/), and PSI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). Transmembrane regions were predicted with TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al., 2001). Phylogenetic tree We selected 31 GPCR amino acid sequences belonging to different families. The extremely variable amino-terminal and carboxy-terminal regions were deleted from all of these receptor sequences. The sequences were then aligned using ClustalW version 1.8. The resulting multiple alignment was used as input to Mega2 (Kumar et al., 2001) to construct a neighbor-joining tree from 1000 replicates of the interior branch test. RT-PCR A panel of total RNA from 20 different human tissues was purchased from Clontech (Palo Alto, CA, USA). Additionally, RNA was prepared from tumor cell lines and tumor tissues by the guanidinium thiocyanate method. RNAs were treated with DNAse (Promega) according to the manufacturer’s instructions. Tumor samples were obtained from the A.C. Camargo Hospital tumor collection. First, 1 µg total RNA, plus 100 ng oligo (dT) in 13.5 µl DEPC-treated water, was incubated for 2 min at 70°C. The reaction was rapidly chilled on ice and used to synthesize cDNA in 20 µl 1X Superscript II first strand buffer containing 0.5 mM dNTP, 3 mM MgCl2, 20 U RNAse inhibitor (RNaseOUT; Invitrogen Life Technologies) and 200 U Superscript II reverse transcriptase at 42°C for 60 min. Control templates for checking amplification of genomic DNA were prepared without reverse transcriptase. The product was diluted to 100 µl and 2.5 µl was used for PCR to amplify BTR. Primers were synthesized by Operon Technologies Inc. The forward and reverse primers match putative transmembrane regions IV and VII, respectively, and should produce a 450-bp long PCR product (BTR1F: 5’ GCTACCTCTCCTTCATGTCC; BTR4R: 5’ GCATCATGAGTACCTCACTG). Twenty-five microliter PCR reactions containing 2.5 µl cDNA, 0.2 mM dNTP, 1.5 mM MgCl2, 0.5 µM of each forward and reverse primers, 1.25 U Platinum Taq DNA polymerase (Invitrogen Life Technologies) were heated to 95°C for 2 min, followed by 35 thermal cycles of 95°C for 45 s, 55°C for 45 s, 72°C for 1 min, and a final incubation at 72°C for 6 min. PCR products were analyzed in 1.5% agarose gels stained with ethidium bromide or in Southern blots using a purified BTR PCR product as a probe. The identity of the PCR products was also confirmed by DNA sequencing. The relative intensities of bands in Figures 2A, 3 and 4 were determined by densitometry using the program LabWorks (UVP). The densities for the different bands were normalized with their corresponding GAPDH band densities. The primers BTRA: 5’ ATGGGGGATGAGCTGGCAC and BTRB: 5’ CTAGGAAATGGTAAAGATGGC were used to amplify the whole coding region of BTR from human brain cDNA. The sequence of the obtained PCR was deposited in Genbank (accession number: AY280965). RESULTS Identification of a novel GPCR In previous experiments we used peptide motifs commonly seen in odorant receptors (OR) as queries to scan the human genome for new OR genes (Malnic et al., 2004). OR genes, like many other GPCR genes, have no introns in their coding sequence (Buck and Axel, 1991), which makes it easier to search for related genes by scanning the human genome databases. Among the many OR gene sequences identified, we found a putative GPCR that did not show any of the common motifs which characterize the OR family members (see Methods). BLASTN and TBLASTN searches, using this translated gene as query, were performed to browse for homologous proteins in the NCBI database (nr and htgs). No highly similar sequences were found: the highest identities were to the human serotonin 1D receptor (5HT1D) (24% AI; 49/202) and alpha-A1-adrenergic receptor (25% AI; 49/190). Although the identity scores to these two GPCRs are very low, a protein motif search with TMHMM (a tool for prediction of transmembrane helices in proteins) indicated seven putative transmembrane regions, suggesting that this gene must code for a new member of the GPCR super family. The fact that we did not find other human proteins closely related to this new putative GPCR indicates that it does not belong to a new family of genes, but instead it is a solitary gene. Chromosomal mapping using Entrez map viewer assigned this gene to chromosome 2q21. During this research the identification of 109 novel human GPCRs out of the human genome sequence (including BTR) was reported by another group (Takeda et al., 2002). A second group also recently reported the identification of 367 total GPCRs in humans, and BTR is also included in this list (Vassilatis et al., 2003). Nevertheless, neither of the two groups described the tissue distribution of BTR. We also scanned the mouse genome sequence from Celera and NCBI (build 30) databases (I.H.G.S.C., 2001; Venter et al., 2001) for a putative homolog for this gene. Neither of the two databases contained sequences related to the BTR gene. We compared the human genomic region containing the human BTR gene (on chromosome 2q21.3) to its corresponding synthenic region on the mouse chromosome 1 (1B). We found mouse orthologs for two human genes flanking the BTR gene in this region, but we found no gene similar to BTR located between these genes. Additionally, no mouse ESTs could be found matching the BTR gene. Thus, it seems that there is no BTR gene in the mouse genome, although we cannot exclude the possibility that gaps in the mouse genome sequences might have precluded us from identifying the mouse counterpart for this gene. Comparison with other GPCRs GPCRs comprise very large and diverse groups of gene families that recognize distinct ligands. Members of the GPCR superfamily can be grouped in different families based on their sequence similarities. The families characterized best so far are: the rhodopsin family (family A), the secretin receptor family (family B), and the metabotropic glutamate receptor family (family C) (Attwood and Findlay, 1994; Kolakowski, 1994; Strader et al., 1995; Bockaert and Pin, 1999; Pierce et al., 2002). In addition to rhodopsin, family A includes the b-adrenergic receptors, serotonin receptors, OR, adenosine receptors, and many others. Family B includes receptors for polypeptide hormones, such as glucagon, secretin and calcitonin. Family C includes all mGluR types, Ca2+ sensing and GABAB receptors, a group of pheromones (termed VRs or G0VN; reviewed in Bargmann, 1997), and a small group of taste receptors, the T1Rs). Other families include the pheromone receptors (VNRs; Dulac and Axel, 1995) and the taste receptors T2Rs (for a review on taste receptors, see Montmayeur and Matsunami, 2002). Sequence identities can be very low among the most distant GPCRs (<20% ASI), but it is expected that receptors with related functions share conserved sequence motifs. In order to determine if BTR can be assigned to one of the known families, we generated a multiple alignment containing 31 GPCR amino acid sequences representing different families and we constructed a phylogenetic tree from 1000 interior branch test replicates. BTR constituted a separate branch, more closely related to family A, although with an interior branch test value of only 38% (Figure 1). This indicates that BTR is not closely related to any of the known families.
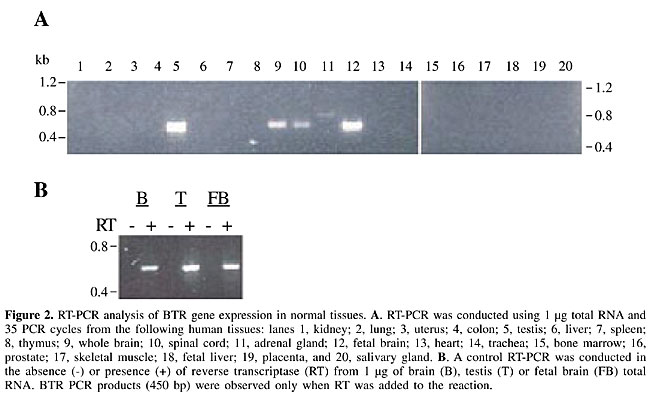
BTR gene expression in normal tissues To investigate the tissue expression pattern of BTR gene, we conducted RT-PCR experiments using a panel of total RNA from 20 different normal human tissues. Expression was detected only in testis, fetal brain, whole brain, and spinal cord (Figure 2A). The gene expression levels in fetal brain, whole brain and spinal cord tissues were detected at 4, 27, and 19%, respectively, of the level detected in the testis (see Methods). When human genomic DNA was used as a template with the same set of primers, a PCR product containing a sequence identical to the cDNA PCR product was obtained, confirming that BTR is an intronless gene. We only detected PCR products when reverse transcriptase (RT) was added to the reaction, which confirmed the absence of contaminating genomic DNA in our RNA samples (Figure 2B). The complete BTR coding region was also amplified from brain cDNA, and the sequence was deposited in Genbank (accession number AY280965). These results indicate that the BTR gene is preferentially expressed in testis and in the nervous system.
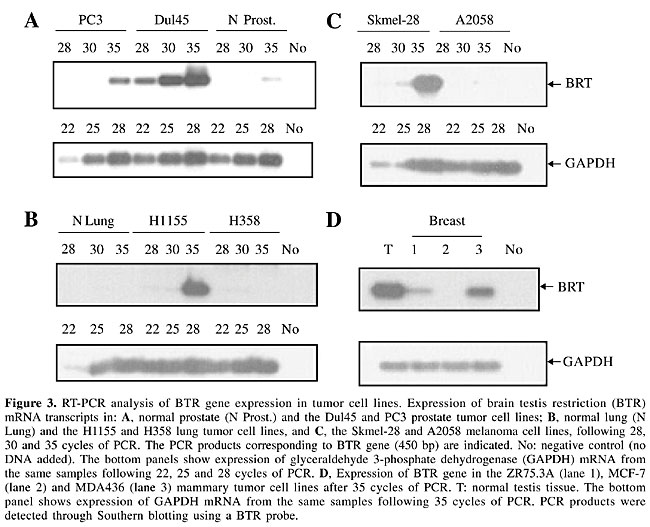
BTR gene expression in tumor cell lines and tissues There are some examples of highly tissue-restricted gene products that are also expressed in cancer. A group of proteins, called cancer/testis/brain antigens (CTB antigens), expressed in normal testis and brain and in different types of tumors has been described. These proteins were originally identified as the target molecules recognized by autoantibodies in patients with paraneoplastic syndromes (Dropcho et al., 1987; Dalmau et al., 1999; Voltz et al., 1999; Scanlan et al., 2002a). We checked whether the BTR gene is also expressed in malignant tissues. We performed RT-PCR using RNA extracted from different cell lines as templates (Figure 3). BTR was expressed in the prostate tumor cell lines Dul45 and PC3, in the H1155 lung tumor cell line, in the Skmel melanoma cell line, and in the MDA436 mammary cell line. We also detected weak BTR gene expression after 35 cycles in normal prostate tissue, which we had not detected before (compare Figures 2A and 3A). This is probably because we used Southern blot to detect the PCR products, which is a more sensitive technique. BTR gene expression in the prostate cell lines was 20X (Dul45) or 7X (PC3) stronger than the expression in normal prostate tissue after 35 cycles.
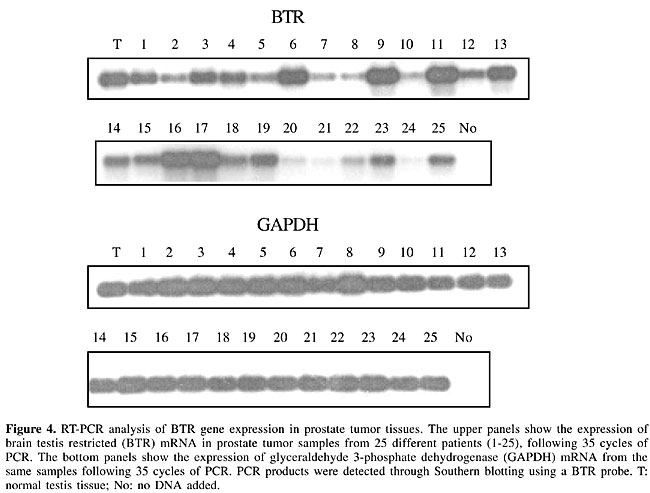
We next assessed whether prostate tumor samples also express the BTR gene. Expression was detected in all prostate tumor samples tested (25/25; Figure 4). Cancer/testis antigen expression may be associated with tumor progression and with tumors of higher malignant potential (Scanlan et al., 2002b); however, this correlation was not observed in our analysis since expression was detected in all samples independently of the progression stage. We compared the relative levels of BTR gene expression in normal prostate tissue and in the prostate tumor samples. Of the 25 samples, only three showed expression levels similar or lower to the one observed for normal prostate tissue (samples 20, 21 and 24). The remaining 22 samples showed higher levels of expression, varying from three to thirty times the amount of BTR expression observed in the normal prostate tissue. These results indicate that BTR gene expression may constitute a consistent marker for prostate cancer.
DISCUSSION GPCRs comprise the largest family of proteins in many species. There are hundreds of GPCRs in humans, many of which have no known ligand or function (called “orphan GPCRs”; Howard et al., 2001). Analysis of the human genome sequence indicates that there are many other unknown GPCRs (I.H.G.S.C., 2001; Venter et al., 2001). We describe the identification and characterization of a novel putative member of this large family of receptors. Comparison of BTR with representative members of the GPCR super family indicates that this novel member constitutes a separate subtype of receptor and may have a quite distinct function, yet to be determined. We found no evidence that there is a mouse BTR gene. It is possible that other GPCRs are also absent in rodents. Indeed, it was previously demonstrated that GPR8, a human orphan GPCR related to opioid and somatostatin receptors, was not found in rodents (Lee et al., 1999). It is believed that most mouse genes have corresponding orthologs in humans, with only a small fraction of the genes (less than 1%) showing no homolog in the other species (Mouse Genome Sequencing Consortium, 2002). These genes must be either involved in specific functions, inherent to one species but not the other, or just be redundant or even non-functional. The identification of all of the species-specific genes through the comparison of the mouse and human genome sequences will clarify if some of these genes play important roles in the determination of characteristics that are specific to human or mouse. The function of the BTR gene product is unknown. The fact that this gene is preferentially expressed in the brain and spinal cord, besides the testis, indicates that its function must be related to physiological aspects of the nervous system. We do not know if this gene is expressed throughout the brain, or if expression is confined to some particular regions within the brain. The identification of the exact regions of BTR expression in the nervous system will probably contribute to the understanding of this receptor’s function. GPCRs have only been recently identified as mediators of cellular growth and differentiation (Dhanasekaran et al., 1995; Luttrell et al., 1999). A large number of studies have also demonstrated that GPCRs can promote tumor formation (for a review, see Whitehead et al., 2001). Recently, a prostate specific GPCR (termed PSGR), which is expressed at higher levels in prostate cancer, was identified (Xu et al., 2000; Xia et al., 2001). Tumor-associated over-expression of PSGR was identified in 62% of the prostate specimens analyzed (32 of 52) (Xu et al., 2000). Like the CTB antigens (Dropcho et al., 1987; Dalmau et al., 1999; Voltz et al., 1999; Scanlan et al., 2002a), the BTR gene is expressed in cancer cell lines and tumors. Whether the BTR gene product is able to elicit an autoimmune response in patients with prostate cancer, still needs to be determined. If this turns out to be the case, the detection of anti-BTR antibodies could be useful for the diagnosis of prostate cancer. In conclusion, we identified a new human GPCR that is expressed preferentially in the normal nervous system and testis. Further studies are required to determine the precise regions within the brain where the BTR gene is expressed, which would contribute to the understanding of BTRs function. We also found a correlation between prostate tumor and elevated levels of BTR gene expression, suggesting a potential role for the gene as a marker for the disease. ACKNOWLEDGMENTS We thank Jean Pierre Montmayeur, Sandro José de Souza and Andrew Simpson for reviewing the manuscript. We also thank the Hospital A.C. Camargo (São Paulo, Brazil) for providing the tumor samples, Dr. Simone Treiger Sredni for help with the description of the clinical stages of the different prostate tumors analyzed in this study and Dr. Ronaldo B. Quaggio for help with the Labworks software. Research supported by FAPESP, by the São Paulo branch of the Ludwig Institute for Cancer Research, and by grants from FAPESP to P.A.F. Galante and from CAPES to R.B. Parmigiani. REFERENCES Attwood, T. and Findlay, J. (1994). Fingerprinting G-protein-coupled receptors. Protein Eng. 7: 195-203. Bargmann, C. (1997). Olfactory receptors, vomeronasal receptors and the organization of olfactory information. Cell 90: 585-587. Bockaert, J. and Pin, J. (1999). Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 18: 1723-1729. Buck, L. and Axel, R. (1991). A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65: 175-187. Dalmau, J., Gultekin, S., Voltz, R., Hoard, R., DesChamps, T., Balmaceda, C., Batchelor, T., Gerstner, E., Eichen, J., Frennier, J., Posner, J. and Rosenfeld, M. (1999). Ma 1, a novel neuron- and testis-specific protein, is recognized by the serum of patients with paraneoplastic neurological disorders. Brain 122: 27-39. Dhanasekaran, N., Heasley, L. and Johnson, G. (1995). G protein-coupled receptor systems involved in cell growth and oncogenesis. Endocrinol. Rev. 16: 259-270. Drews, J. (2000). Drug discovery: a historical perspective. Science 287: 1960-1964. Dropcho, E., Chen, Y., Posner, J. and Old, L. (1987). Cloning of a brain protein identified by autoantibodies from a patient with paraneoplastic cerebellar degeneration. Proc. Natl. Acad. Sci. USA 84: 4552-4556. Dulac, C. and Axel, R. (1995). A novel family of genes encoding putative pheromone receptors. Cell 83: 195-206. Howard, A., McAllister, G., Feighner, S., Liu, Q., Nargund, R., Van de Ploeg, L. and Patchett, A. (2001). Orphan G protein-coupled receptors and natural ligand discovery. Trends in Pharmacol. Sci. 22: 132-140. I.H.G.S.C. (2001). Initial sequencing and analysis of the human genome. Nature 409: 860-921. Kolakowski, L.F. (1994). GCRDb: a G-protein-coupled receptor database. Receptors Channels 2: 1-7. Krogh, A., Larsson, B., Von Heijne, G. and Sonnhammer, E. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567-580. Kumar, S., Tamura, K., Jakobsen, I. and Nei, M. (2001). MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244-1245. Lee, D., Nguyen, T., Porter, C., Cheng, R., George, S. and O’Dowd, B. (1999). Two related G protein-coupled receptors: the distribution of GPR7 in rat brain and the absence of GPR8 in rodents. Mol. Brain Res. 71: 96-103. Lee, D., George, S. and O’Dowd, B. (2002). Novel G protein-coupled receptor genes expressed in the brain: continued discovery of important therapeutic targets. Expert Opin. Ther. Targets 6: 185-202. Luttrell, L., Daaka, Y. and Lefkowitz, R. (1999). Regulation of tyrosine kinase cascades by G-protein coupled receptors. Curr. Opin. Cell Biol. 11: 177-183. Malnic, B., Godfrey, P. and Buck, L. (2004). The human olfactory receptor gene family. Proc. Natl. Acad. Sci. 101: 2584-2589. Marchese, A., George, S., Kolakowski Jr., L., Lynch, K. and O’Dowd, B. (1999). Novel GPCRs and their endogenous ligand: expanding the boundaries of physiology and pharmacology. Trends Pharmacol. Sci. 20: 370-375. Montmayeur, J. and Matsunami, H. (2002). Receptors for bitter and sweet taste. Curr. Opin. Neurobiol. 10: 519-527. Mouse Genome Sequencing Consortium (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. Pierce, K., Premont, R. and Lefkowitz, R. (2002). Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3: 639-650. Scanlan, M., Gordon, C., Williamson, B., Lee, S., Chen, Y., Stockert, E., Jungbluth, A., Ritter, G., Jager, D., Jager, E., Knuth, A. and Old, L. (2002a). Identification of cancer/testis genes by database mining and mRNA expression analysis. Int. J. Cancer 98: 485-492. Scanlan, M., Gure, A., Jungbluth, A., Old, L. and Chen, Y. (2002b). Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol. Rev. 188: 22-32. Strader, C., Ming Fong, T., Graziano, M. and Tota, M. (1995). The family of G-protein-coupled receptors. FASEB J. 9: 745-754. Takeda, S., Kadowaki, S., Haga, T., Takaesu, H. and Mitaku, S. (2002). Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Let. 520: 97-101. Vassilatis, D., Hohmann, J., Zeng, H., Li, F., Ranchalis, J., Mortrud, M., Brown, A., Rodriguez, S., Weller, J., Wright, A., Bergmann, J. and Gaitanaris, G. (2003). The G protein-coupled receptor repertoires of human and mouse. Proc. Natl. Acad. Sci. USA 100: 4903-4908. Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M. and Evans, C.A. et al. (2001). The sequence of the human genome. Science 291: 1304-1351. Voltz, R., Gultekin, S., Rosenfeld, M., Gerstner, E., Eichen, J., Posner, J. and Dalmau, J. (1999). A serologic marker of paraneoplastic limbic and brain-stem encephalitis in patients with testicular cancer. N. Engl. J. Med. 340: 1788-1795. Whitehead, I., Zohn, I. and Der, C. (2001). Rho GTPase-dependent transformation by G protein-coupled receptors. Oncogene 20: 1547-1555. Xia, C., Ma, W., Wang, F., Hua, S.-B. and Liu, M. (2001). Identification of a prostate-specific G-protein coupled receptor in prostate cancer. Oncogene 20: 5903-5907. Xu, L., Stackhouse, B., Florence, K., Zhang, W., Shanmugam, N., Sesterhenn, I., Zou, Z., Srikantan, V., Augustus, M., Roschke, V., Carter, K., McLeod, D., Moul, J., Soppett, D. and Srivastava, S. (2000). PSGR, a novel prostate-specific gene with homology to a G protein-coupled receptor, is overexpressed in prostate cancer. Cancer Res. 60: 6568-6572. |
|