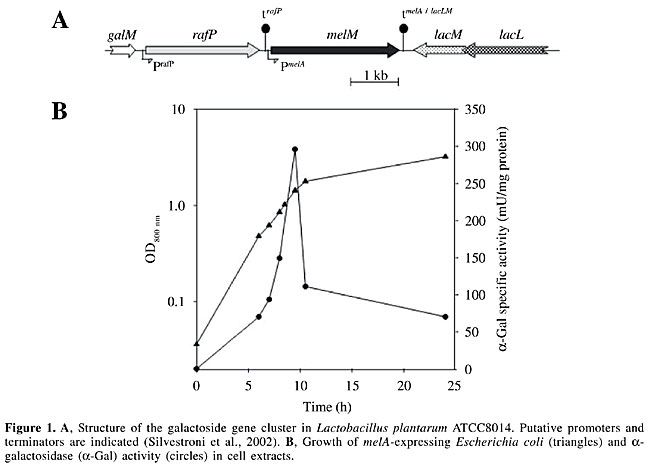
ABSTRACT. Human consumption of soy-derived products has been limited by the presence of non-digestible oligosaccharides (NDO), such as the a-galactooligosaccharides raffinose and stachyose. Most mammals, including man, lack pancreatic a-galactosidase (a-Gal), which is necessary for the hydrolysis of these sugars. However, such NDO can be fermented by gas-producing microorganisms present in the cecum and large intestine, which in turn can induce flatulence and other gastrointestinal disorders in sensitive individuals.The use of microorganisms expressing a-Gal is a promising solution to the elimination of NDO before they reach the large intestine. In the present study, lactic acid bacteria engineered to degrade NDO have been constructed and are being used as a tool to evaluate this solution. The a-Gal structural genes from Lactobacillus plantarum ATCC8014 (previously characterized in our laboratory) and from guar have been cloned and expressed in Lactococcus lactis. The gene products were directed to different bacterial compartments to optimize their possible applications. The a-Gal-producing strains are being evaluated for their efficiency in degrading raffinose and stachyose: i) in soymilk fermentation when used as starters and ii) in situ in the upper gastrointestinal tract when administered to animals orally, as probiotic preparations. The expected outcomes and possible complications of this project are discussed. Key words: Alpha-galactosidase, Genetically modified microorganisms, Lactic acid bacteria, Alpha-galactosides, Soy INTRODUCTION Among protein sources, soy products have an excellent nutritional status based on their high protein content and soy proteins contain enough of all the essential amino acids to meet biological requirements when consumed at the recommended level of protein intake. However, the increase in human consumption of soy products has been limited due to the presence of non-digestible oligosaccharides (NDO, i.e., a-galactosides such as raffinose and stachyose) in soybeans, which are not eliminated by usual soy processing (Leske et al., 1993). NDO have been defined as “carbohydrates with a degree of polymerization of two or more, which are soluble in 80% ethanol and are not susceptible to digestion by pancreatic and brush border enzymes” (Quigley et al., 1999). In soybeans, a-galactosides represent 4-6% of the dry mass. Their hydrolytic digestion in the small intestine is relatively weak because most mammals do not express pancreatic a-galactosidase (a-Gal) necessary to hydrolyze the a-1,6 linkages of these sugars (Slominski, 1994). In humans, these NDO are metabolized by microorganisms in the large intestine, liberating huge amounts of gas that can cause gastrointestinal disorders in sensitive individuals. Moreover, they have been reported to increase the incidence of diarrhea in rats (Kuriyama and Mendel, 1917). Although the use of microbial a-Gal appears to be a promising solution for the degradation of these NDO in soy products, dietary supplementation with a-Gal has not yet demonstrated conclusive benefits. Recently, Smiricky et al. (2002) showed only little improvement in the digestibility of NDO in soy products that were supplemented with high levels of a-Gal and fed to swine. These authors suggested that the enzyme might have been denatured or inactivated when it reached the hostile conditions of the stomach. In this article, the use of engineered lactic acid bacteria (LAB) to degrade NDO is proposed. The constructed a-Gal-producing strains are being evaluated for their efficiency in degrading raffinose and stachyose: i) in soymilk fermentation when used as starters and ii) in situ in the upper gastrointestinal tract when administered orally to animals as probiotic preparations. The expected outcomes and possible complications of this project are discussed. CHOICE OF MODEL STRAINS LAB present in fermented foods have long been consumed by humans without any obvious adverse effects (Fuller, 1992). Therefore, they are potent candidates as vehicles for the delivery of digestive enzymes. LAB, including some Lactobacillus (Lb.) plantarum, Lb. fermentum, Lb. brevis, Lb. buchneri, and Lb. reuteri strains, hydrolyze a-galactosides into digestible carbohydrates during vegetable fermentation (Cruz et al., 1981). These bacteria are therefore a source of a-Gals. The choice of the model strains to be used in our study was made according to the following rationale. Although Lactococcus lactis does not produce a-Gal, this bacterium is by far the most extensively studied LAB, and over the past decades, a number of elegant and efficient genetic tools have been developed for this starter bacterium. These tools are of critical importance in metabolic engineering strategies that aim to inactivate undesired genes and/or to control the expression of existing or novel ones. The nisin-controlled expression system for controlled heterologous and homologous gene expression in L. lactis is a fine example of emerging strategies that can be used to improve the potential applications of LAB (de Ruyter et al., 1996). The expression of heterologous proteins and antigens, as well as the various delivery systems developed in L. lactis, have recently been reviewed (Nouaille et al., 2003). The ability of L. lactis to survive the hostile conditions of the human gastrointestinal tract has made it the model bacterium for the delivery of gene products in the gut (Klijn et al., 1995). Drouault et al. (1999) have extensively studied the survival, physiology, and lysis of this LAB in the digestive tract. The results of their studies show that lactococci are quite resistant to gastric acidity (98% survival) but only 10-30% survive in the duodenum. Viable cells are metabolically active in each compartment of the digestive tract, whereas most dead cells appear to be subject to rapid lysis. These properties suggest that L. lactis could be used as a vector to specifically deliver proteins produced in the cytoplasm into the duodenum of monogastric animals. Together, these features led us to choose L. lactis as the bacterial vehicle for a-Gal in this study. CHOICE OF a-GALACTOSIDASE GENES Recently, we reported the molecular characterization of an a-Gal from Lb. plantarum (Silvestroni et al., 2002). This LAB has been widely studied because it is found in the natural microbiota of healthy humans (Kleerebezem et al., 2003). The a-Gal-encoding melA gene of Lb. plantarum ATCC8014 was probed using polymerase chain reaction (PCR) with degenerate primers and the gene cluster was characterized using uneven and inverse PCR techniques. This allowed us to unravel the structure of the galactoside-metabolism gene cluster shown in Figure 1A. The melA gene encodes a 738-amino acid protein (MelA) with a deduced molecular mass of 84 kDa. The melA gene is surrounded by genes involved in a- and b-galactoside utilization. The galM gene encodes a putative mutarotase, rafP a putative raffinose transporter with high homology to lacS, the bifunctional lactose and a-galactoside transporter from Streptococcus thermophilus, and lacL and lacM, the two putative subunits of b-galactosidase. In silico analysis of the melA gene product did not indicate the presence of sorting signals, suggesting that this a-Gal occurs as a soluble enzyme in the cytoplasm of Lb. plantarum. This has been confirmed by fractionated extraction experiments. Molecular weight assessment of active MelA showed that it occurs as oligomers and that the monomers are inactive. Northern hybridization revealed that melA is expressed in Lb. plantarum ATCC8014 and that it is transcribed from its own promoter. Regulation occurs at the transcriptional level, i.e., melA is induced by the a-galactoside melibiose. The melA gene was expressed in Escherichia coli RA11r, a melA-negative strain, yielding active MelA (Figure 1B).
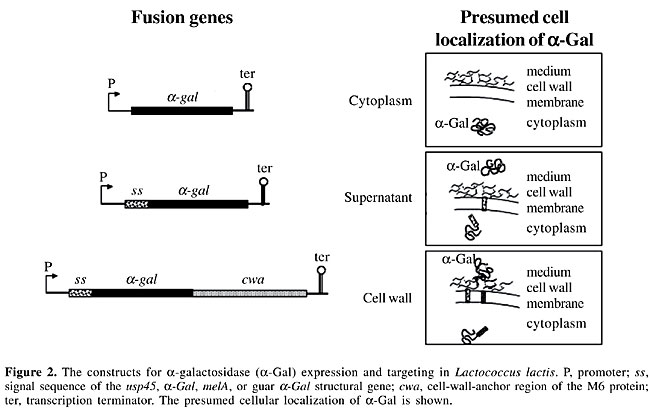
The a-Gal gene of the legume Cyamopsis tetragonoloba (guar), encoding a 40-kDa monomeric protein, has previously been reported (Overbeeke et al., 1989). This protein is active on the chromogenic substrate 5-bromo-4-chloro-3-indolyl a-D-galactopyranoside (X-a-Gal), the trisaccharide raffinose, and the galactomannan substrate guar gum. Each a-Gal offers different advantages and disadvantages, which is described in the following sections. GENETIC ENGINEERING IN LACTOBACILLUS LACTIS The a-Gal structural genes from Lb. plantarum and from guar were inserted into the different L. lactis vectors shown in Figure 2. The gene products (a-Gal) were directed to different bacterial compartments using an expression and export system designed to allow the targeting of gene products to specific locations in L. lactis cells (i.e., cytoplasm, cell wall, or medium) to optimize their possible applications (Dieye et al., 2001). These vectors contain the strong constitutive P59 promoter or the nisin-inducible Pnis promoter of L. lactis (van der Vossen et al., 1987; de Ruyter et al., 1996), the signal sequence of Usp45 for the membrane translocation of secreted and cell-wall-anchored a-Gals (van Aeeldonk et al., 1990), and the cell-wall-anchor domain of the M6 protein for cell-surface display of a-Gals by the sortase machinery (Hollingshead et al., 1986; Mazmanian et al., 1999).
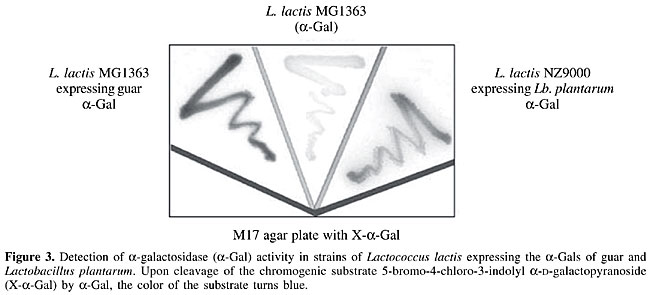
The expression of both a-Gal genes in L. lactis was assessed on M17-agar plates containing the chromogenic substrate X-a-Gal (Figure 3). Specific antibodies directed against the a-Gals of Lb. plantarum and guar are currently being produced. These antibodies are to be used in Western analyses of fractioned protein extracts to confirm that the bacterial localizations of the a-Gals are correct.
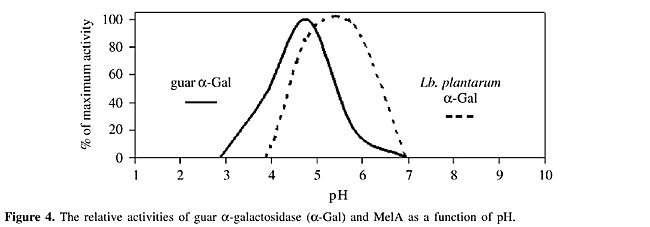
SOYMILK FERMENTATION In soymilk fermentation, the degradation of a-galactosides and the growth of the engineered strains are monitored throughout the fermentation process. In these studies, the pH constraint is very important, because the initial pH (near 7.6) is close to neutrality but steadily decreases during fermentation due to the conversion of sugars into organic acids by the starter cultures. To address this constraint, L. lactis expressing a-Gals with different ranges of optimal pH are used. The a-Gal activities of Lb. plantarum and guar differ significantly in their functional pHs, the former being most active between pH 4.5 and 5.2 and the latter between pH 5.0 and 6.2 (Figure 4).
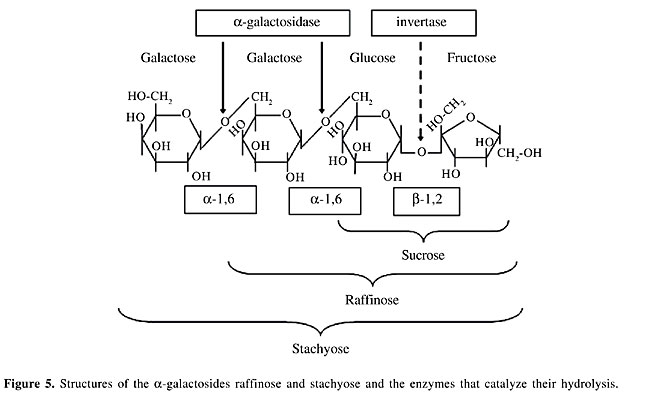
Another important constraint to consider in soymilk fermentation is substrate accessibility. The construction of different expression and export vectors allows the a-Gal to be secreted into the medium (soymilk) or to be located in the cytoplasm or cell wall of L. lactis. The excreted form would be optimal because the a-Gals are transported outside the bacteria. This allows the enzyme to be in direct contact with its substrates. A drawback of this localization is that the secreted enzyme is exposed to the environmental conditions of the fermentation vessel, such as pH, salts, and denaturing agents. On the other hand, although cytoplasmic localization would protect the enzyme, it requires the presence of an efficient functional a-galactoside transporter. Halfway between these two localization options is cell-wall anchoring of a-Gals, which could favor substrate-enzyme interactions while the cell-wall microenvironment would have a partial protective effect on enzyme stability. ANIMAL STUDIES The constructed strains are being tested in vivo using an animal model. In these studies, rodents fed soy-based diets receive, by oral administration, live a-Gal-producing strains. The ability of these engineered strains to reduce non-digestible sugars is determined using different parameters. First, the a-Gal activity is determined in each compartment of the gastrointestinal tract, such as the stomach, duodenum, jejunum, ileum, cecum, and large intestine. An increase in a-Gal activity before the cecum is the goal because this would permit NDO such as raffinose and stachyose to be degraded into smaller compounds (sucrose and galactose, see Figure 5) that are normally absorbed in the small intestine. If this is the case, the a-Gal activity of the cecum and the overall size of this organ would decrease because of a diminution of the fermentative activities of microorganisms stimulated by the arrival of oligosaccharides in this intestinal compartment (LeBlanc et al., 2004).
Second, the degradation of a-galactosides in situ is being evaluated. Here, the residual sugar content of each intestinal compartment is determined by high-performance liquid chromatography. Finally, gas production is monitored. Hydrogen, carbon dioxide, and sometimes methane gases are produced by the bacterial fermentation of unabsorbed carbohydrates in the large intestine. The level of production of these gases can be used as an index of the amount of unabsorbed carbohydrate that passes through the small intestine. In in vivo trials, the strains producing a-Gals of different origins and expressed in different cellular localizations are evaluated comparatively to see which exhibits the most appropriate response to the hostile conditions of the gastrointestinal tract. Here, the secreted form is probably the least suitable because the a-Gal produced would be in the presence of digestive enzymes, salts, extreme pH conditions, etc. The cytoplasmic form is a very promising option because L. lactis is lysed in the duodenum and an a-Gal-overproducing strain would therefore be able to liberate large quantities of this enzyme in the upper intestinal tract. However, the liberated enzyme must be quick-acting, because it would then be exposed to the harsh conditions described above. As in the fermentation studies, cell-wall anchoring of a-Gals should favor substrate-enzyme interactions, while the cell-wall microenvironment would have a partial protective effect on enzyme stability. OTHER STUDIES The search for natural LABs exhibiting the required phenotypes in terms of their levels of a-Gal activity is an alternative strategy. However, most a-Gal-positive LABs produce this enzyme in the cytoplasm, limiting their possible use. We have also observed that strains with the highest a-Gal activities are often unsuitable for in vivo uses (LeBlanc, J.G., Sesma, F. and Savoy de Giori, G., unpublished data). Poor enzyme stability, low survival rates in the gastrointestinal tract, and secondary effects are factors that strongly limit the applicability of most microorganisms available for animal use. The genetic manipulation of strains, as in this study, can improve the functions of LABs in producing desired effects not found in natural strains. EXPECTED OUTCOMES OF THE PROJECT The improvement of LAB functions is of great importance for existing food manufacturing processes, so that the final product better suits the digestive capacities of consumers. During the past decades, much work has improved the technological output of LABs. We are now on the road to improving their performance in a nutritional context. In terms of the removal of a-galactosides from soy-derived products, various methods (soy germination, bean soaking, and water extraction) exist, but all are laborious. Obviously, the utilization of LAB strains that perform both fermentation and a-galactoside removal is economically attractive. It is also very important to demonstrate that the probiotic activity of LABs is unambiguous. To achieve this, we chose to overexpress an enzymatic activity (a-Gal) that is deficient in man and in L. lactis and to show that this activity occurs in vivo, using an animal model. Both of these issues may contribute to the entry of LABs into the field of nutraceuticals. Other issues addressed by the European Nutra Cells consortium (www.nutracells.com), which contains the present project, include vitamin production by LABs, production of the non-metabolized sugars mannose and trehalose by LABs, removal of lactose and galactose from dairy products, and oligosaccharide production using LABs. These achievements should open the door to many applications in the development of both new food products with enhanced nutritional value and probiotic preparations with well-demonstrated in vivo activity. The ultimate use of such bacteria may rely on the still controversial decision about the acceptability of genetically modified organisms in nutrition and nutraceutical preparations. Undoubtedly, the consumers will be the major players in this decision. The scientifically proven health benefits that can be gained by consumption of these genetically modified organisms are clearly one of the elements in this decision. ACKNOWLEDGMENTS Research supported by contract QLK1-CT-2000-01376 of the EEC (http://www.nutracells.com). We are grateful to Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Agencia Nacional de Promoción Científica y Tecnológica (ANPyCT-FONCYT) (Argentina) and Ecos-Sud (France) for their support of this project, to Lallemand for its interest and collaboration in this study, and to Dr. M. Garro and F. Clier for their valuable expertise in the course of our studies. REFERENCES Cruz, R., Bastistela, J.C. and Wosiacki, G. (1981). Microbial a-galactosidase for soybean processing. J. Food Sci. 46: 1196-1200. de Ruyter, P.G., Kuipers, O.P. and de Vos, W.M. (1996). Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62: 3662-3667. Dieye, Y., Usai, S., Clier, F., Gruss, A. and Piard, J.C. (2001). Design of a protein-targeting system for lactic acid bacteria. J. Bacteriol. 83: 4157-4166. Drouault, S., Corthier, G., Ehrlich, D. and Renault, P. (1999). Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Appl. Environ. Microbiol. 65: 4881-4886. Fuller, R. (1992). History and development of probiotics. In: Probiotics - The Scientific Basis (Fuller, R., ed.). Chapman and Hall, London, England, pp. 1-8. Hollingshead, S.K., Fischetti, V.A. and Scott, J.R. (1986). Complete nucleotide sequence of type 6 M protein of the group A Streptococcus. Repetitive structure and membrane anchor. J. Biol. Chem. 261: 1677-1686. Kleerebezem, M., Boekhorst, J., van Kranenburg, R., Molenaar, D., Kuipers, O.P., Leer, R., Tarchini, R., Peters, S.A., Sandbrink, H.M., Fiers, M.W., Stiekema, W., Lankhorst, R.M., Bron, P.A., Hoffer, S.M., Groot, M.N., Kerkhoven, R., de Vries, M., Ursing, B., de Vos, W.M. and Siezen, R.J. (2003). Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100: 1990-1995. Klijn, N., Weerkamp, A.H. and de Vos, W.M. (1995). Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl. Environ. Microbiol. 61: 2771-2774. Kuriyama, S. and Mendel, L.B. (1917). The physiological behavior of raffinose. J. Biol. Chem. 31: 125-147. LeBlanc, J.G., Garro, M.S., Silvestroni, A., Connes, C., Piard, J.-C., Sesma, F. and Savoy de Giori, G. (2004). Reduction of a-Galactooligosaccharides in Soyamilk by Lactobacillus fermentum CRL 722: In vitro and in vivo evaluation of fermented soyamilk. J. Appl. Microbiol. 97: 876-881. Leske, K.L., Jevne, C.J. and Coon, C.N. (1993). Effect of oligosaccharide additions on nitrogen-corrected true metabolizable energy of soy protein concentrate. Poult. Sci. 72: 664-668. Mazmanian, S.K., Liu, G., Ton-That, H. and Schneewind, O. (1999). Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285: 760-763. Nouaille, S., Ribeiro, L.A., Miyoshi, A., Pontes, D., Le Loir, Y., Oliveira, S.C., Langella, P. and Azevedo, V. (2003). Heterologous protein production and delivery sytems for Lactococcus lactis. Genet. Mol. Res. 2: 102-111 (www.funpecrp.com.br/GMR). Overbeeke, N., Fellinger, A.J., Toonen, M.Y., van Wassenaar, D. and Verrips, C.T. (1989). Cloning and nucleotide sequence of the alpha-galactosidase cDNA from Cyamopsis tetragonoloba (guar). Plant Mol. Biol. 13: 541-550. Quigley, M.E., Hudson, G.J. and Englyst, H.N. (1999). Determination of resistant short-chain carbohydrates (non-digestible oligosaccharides) using gas-liquid chromatography. Food Chem. 65: 381-390. Silvestroni, A., Connes, C., Sesma, F., Savoy de Giori, G. and Piard, J.C. (2002). Characterization of the melA locus for alpha-galactosidase in Lactobacillus plantarum. Appl. Environ. Microbiol. 68: 5464-5471. Slominski, B.A. (1994). Hydrolysis of galactooligosaccharides by commercial preparations of alpha-galactosidase and beta-fructofuranose: Potential for use as dietary additives. J. Sci. Food Agric. 65: 323-330. Smiricky, M.R., Grieshop, C.M., Albin, D.M., Wubben, J.E., Gabert, V.M. and Fahey Jr., G.C. (2002). The influence of soy oligosaccharides on apparent and true ileal amino acid digestibilities and fecal consistency in growing pigs. J. Anim. Sci. 80: 2433-2441. van Aeeldonk, M., Rutten, G., Oteman, M., Siezen, R.J., de Vos, W.M. and Simons, G. (1990). Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. Lactis MG1363. Gene 25: 155-160. van der Vossen, J.M., van der Lelie, D. and Venema, G. (1987). Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl. Environ. Microbiol. 53: 2452-2457. |
|