ABSTRACT. Virus-induced gene silencing (VIGS) has been shown to be of great potential in plant reverse genetics. Advantages of VIGS over other approaches, such as T-DNA or transposon tagging, include the circumvention of plant transformation, methodological simplicity and robustness, and speedy results. These features make VIGS an attractive alternative instrument in functional genomics, even in a high throughput fashion. The system is already well established in Nicotiana benthamiana; however, efforts are being made to improve VIGS in other species, including monocots. Current research is focussed on unravelling the mechanisms of post-transcriptional gene silencing and VIGS, as well as on finding novel viral vectors in order to broaden the host species spectrum. We examined how VIGS has been used to assess gene functions in plants, including molecular mechanisms involved in the process, available methodological elements, such as vectors and inoculation procedures, and we looked for examples in which the system has been applied successfully to characterize gene function in plants. Key words: Nicotiana benthamiana, Reverse genetics, Post-transcriptional gene silencing INTRODUCTION Reverse genetics is the search for gene functionality, starting from a gene sequence. Knocking out genes is the most frequently used strategy of reverse genetics to infer about gene functions. Two insertional mutagenesis approaches for gene disruption have been predominantly used in Arabidopsis: transferred DNA (T-DNA) (Krysan et al., 1999) and transposon tagging (Parinov et al., 1999; Speulman et al., 1999). Although these are powerful tools for providing novel mutants, they present some limitations, including the impossibility of studying the function of duplicated genes, the difficulty to reach genome saturation, and the multiple insertional nature of these approaches, which frequently leads to concomitant disruption of several genes. The use of tissue-specific promoters with antisense RNA (asRNA) and co-suppression technologies may prevent embryonic developmental obstacles but they are also labor intensive, time consuming, and, in some cases, unpredictable. These constraints can be circumvented by new post-transcriptional gene silencing (PTGS) tools, through which genes are silenced in a specific and efficient manner, using less intensive and time-consuming technologies than by conventional techniques (Wang and Wagner, 2003). PTGS (Covey et al., 1997; Ratcliff et al., 1997; Al-Kaff et al., 1998) can be induced in plants by viral vectors harboring specific genes, through the virus-induced gene silencing (VIGS) system (Kumagai et al., 1995; Baulcombe, 1999), by inverted repeat transgenes producing hairpin transcripts (hairpin RNAs, hpRNAs) (Smith et al., 2000), by asRNA technology (Rothstein et al., 1987), or by gene overexpression (co-suppression) (Napoli et al., 1990; van der Krol et al., 1990). The VIGS system can be very helpful to assess gene function, especially in species recalcitrant to transformation. We examined the advantages of and current knowledge on genetics and biochemistry of PTGS and VIGS. In addition, we studied molecular flower development in order to explore the possibility of using VIGS in model species for probing flower gene functions in heterologous species. ADVANTAGES AND APPLICATIONS OF GENE SILENCING PLANTS Since the great majority of plant virus genomes consist of single-stranded RNA (ssRNA), gene delivery in the VIGS system is often performed with the inoculation of viral vector transcripts synthesized in vitro. In the case of DNA viruses, such as geminivirus, inoculation is simplified because it requires only the corresponding viral DNA. Alternatively, the virus genome can also be inserted as a cDNA fragment into a binary vector, to be introduced into the plant cell via agroinfection, facilitating the delivery process. Genes involved in early developmental stages usually cannot be ascertained by gene disruption approaches, such as T-DNA and transposon tagging, because their lack of function leads to plant death. With the VIGS system, these genes can be silenced after the critical point of germination, to elicit specific silencing from this moment on, in order to understand their post-embryonic developmental roles (Peele et al., 2001). In the genomics age, molecular biologists are looking for new alternatives to study gene function on a genome-wide scale. The high throughput techniques for gene discovery and expression analysis, such as whole genome sequencing and micro-arrays, demand efficient procedures to unravel gene functions, in order to make them useful for fundamental and applied purposes. The information generated by these technologies can be combined with PTGS approaches to probe gene function in a high throughput fashion. This is already a reality in developmental genetics of the nematode Caenorhabditis elegans (Maeda et al., 2001). Traditional gene knockout techniques use transformation as a delivery system, and they usually require tissue culture steps to regenerate silenced mutants. PTGS, and especially the VIGS system, can potentially be used as an important tool in reverse genetic analysis, to ease and speed up analysis of gene function without the requirement of time-consuming transformation and tissue culture procedures, since it can be applied in ex vitro developing plants, switching off genes specifically and in an efficient way, and allowing rapid monitoring. Scientists who study the genes involved in plant defence against pathogens and in cell development, such as transcription factors, are now using the VIGS system to facilitate functional analysis of plant genes. Large-scale loss-of-function studies can now be performed using specific vectors and highly efficient cloning systems, such as the Gateway recombination-based system, to insert gene fragments in a more efficient way (Liu et al., 2002a). The resulting phenotypes can be evaluated within days after inoculation, instead of months, or even years, when the traditional methods that require transformation procedures are used. It is important, however, to take into consideration that until now this system has been efficiently used in only a few plant species, such as Nicotiana benthamiana, tomato, and barley. Nevertheless, considerable efforts are being put into developing novel vectors in order to increase the number of species that respond efficiently to VIGS. Summing up, the VIGS system can be used in reverse genetics to ease and speed up functional analysis, without gene disruption, without requiring plant transformation, and allowing the study of multiple genes concomitantly. BRIEF HISTORY OF GENE SILENCING PTGS is a general term that encompasses similar events occurring in diverse biological research fields. Plant biologists call it PTGS and co-suppression, whereas RNA interference (RNAi) is the term originally used by those working with C. elegans and Drosophila, and quelling is used by fungi scientists (Cogoni and Macino, 2000). Since these phenomena have very similar mechanisms, RNAi is being now used more frequently as the general term to describe the event in other kingdoms as well. asRNA technology was discovered in plant research in 1987, with the inhibition of a nopaline synthase gene in tobacco cells, due to the expression of its corresponding asRNA (Rothstein et al., 1987). The first reports on gene silencing induced by co-suppression were published in 1990 (Napoli et al., 1990; van der Krol et al., 1990) when scientists were trying to increase the purple pigmentation of petunia petals by sense overexpression of CHALCONE SYNTHASE (CHS), a gene related to the anthocyanin pathway. However, this approach induced variegated and white petals, caused by silencing of the endogenous chalcone synthase gene. Interestingly, the white color of the petals was observed in subsequent generations, and occasionally some plants reverted to purple, indicating that the event did not involve permanent DNA modification. This phenomenon was then designated co-suppression, and it intrigued the scientific community for a decade. Similar events were observed in the fungus Neurospora crassa, when Cogoni et al. (1994) attempted to increase its orange pigmentation by introducing a gene involved in carotenoid production. Instead of turning deep orange, the fungus sometimes bleached out. They denominated this process, quelling. The bleached fungus frequently reverted to normal color, indicating that the quelling process did not permanently alter the DNA. Guo and Kemphues (1995) showed the first evidence that sense RNA could instigate gene silencing in C. elegans. Intending to use asRNA to silence a gene that regulates embryo symmetry, they also observed gene silencing when they used the sense RNA as a control. Fire et al. (1998), also working with C. elegans, found that although single-stranded asRNA could trigger gene silencing, double-stranded RNA (dsRNA) was much more effective. They coined the term RNAi for this process. The VIGS system was developed from studies on the sequence homology between a virus and either a transgene or an endogenous gene that would cause PTGS (Lindbo et al., 1993; Kumagai et al., 1995). In this system, a virus vector carrying a copy of the gene to be silenced is introduced into the cell, the cellular machinery recognizes the viral threat and releases a protective defence to destroy not only viral genes but also any extra-gene being carried by the viral vector, affecting any native or transgenic homologous transcripts (Ruiz et al., 1998; Waterhouse et al., 2001). The first cues on the molecular mechanisms underlying PTGS were derived from studies in plants, with a class of small RNAs of about 25 nucleotide (nt) identified as the triggering signal for gene silencing (Hamilton and Baulcombe, 1999). Genetic and molecular studies have confirmed that co-suppression, RNAi, quelling, and VIGS share similar mechanisms of gene silencing that are widely present in eukaryotic species (Gura, 2000). CURRENT MECHANISM OF PTGS IN PLANTS Characterization of the PTGS mechanism is still in its infancy. Much effort is being made to elucidate the genes and biochemistry involved in the defensive response, since it affects gene silencing. The mechanisms seem to be conserved throughout evolution since homologous genes involved in the process have been found in various species of fungi and plants, as well as in animals. The present model for gene silencing (Figure 1) includes three phases: initiation, maintenance, and signal amplification and dissemination (Nishikura, 2001). dsRNA is the triggering factor for gene silencing, and the first phase involves its synthesis or formation, its recognition, and the production of small interfering RNA (siRNAs) fragments. dsRNAs can be generated by the RNA virus replication mechanism, which includes the formation of dsRNA by an RNA-dependent RNA polymerase (RdRP), by hpRNA, originated from a bidirectionally cloned transgene or by an asRNA cloning strategy. Still in the initiation phase, the next step involves mRNA targeting and degradation by the enzyme Dicer (Bernstein et al., 2001), which recognizes and cleaves dsRNAs from both ends into siRNAs of 21 to 23 nt (Zamore et al., 2000). 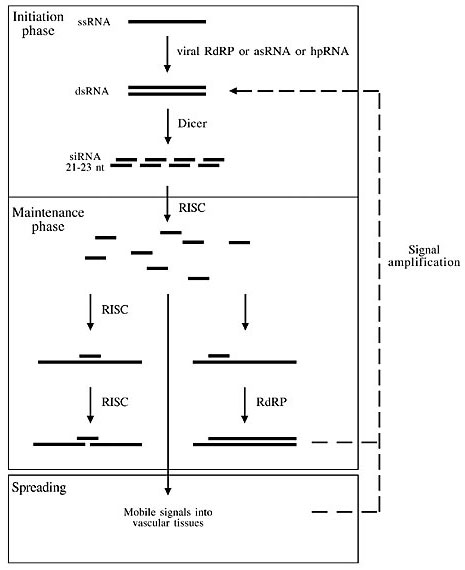
Figure 1. Current model of post-transcriptional gene silencing. The initiation phase can be triggered by a viral RNA-directed RNA polymerase (RdRP) that makes a complementary RNA strand, using the single-stranded as a template, by a transgenic antisense RNA (asRNA ) or by a hairpin RNA (hpRNA). Double stranded RNA (dsRNA) is recognized by the Dicer enzyme and is chopped into pieces of 21-23 nucleotides, called small interfering RNAs (siRNAs). In the maintenance phase, RNA-induced silencing complex (RISC) unwinds the siRNAs, triggers the surveillance mechanism to find siRNA complementary RNA in the cell, and inactivates them with its RNase activity. Alternatively, siRNAs can serve as primers for an RdRP to make double-stranded RNA during the silencing signal amplification, in a feedback fashion. Moreover, siRNAs are believed to constitute (at least partially) mobile signals that spread the gene silencing mechanism to other parts of the plant, through vascular tissues. ssRNA = single-stranded RNA.
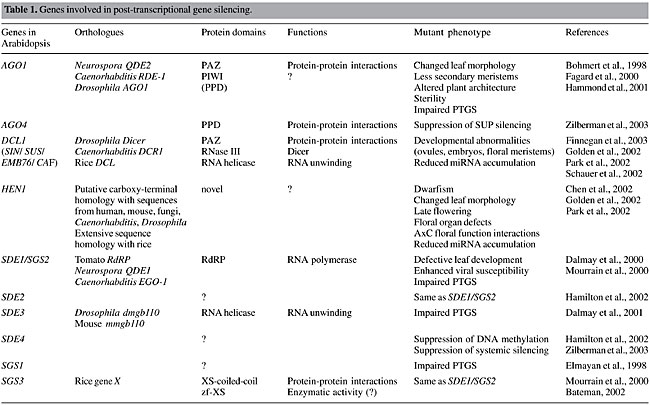
These siRNAs, alternatively referred to as guide RNAs, are identification sequences for the RNA-induced silencing complex (RISC), a protein-RNA effector nuclease formation of about 500 kDa, in the maintenance phase (Hammond et al., 2000). RISC has exo- and endonuclease activities, an RNA homology-searching activity and a helicase to unwind the dsRNA. This complex can be activated by unwinding siRNAs, in order to use their single-stranded siRNA sequences for identification and degradation of complementary transcripts (Nykänen et al., 2001). The third phase concerns the amplification of a silencing signal, and its dissemination throughout the target transcript. This signal is being identified as the siRNAs originated in the preceding phases, and it is alternatively referred to as transitive RNAi. Guided by an RdRP, in order to amplify and potentiate the silencing response, siRNAs act as activators (primers) for dsRNA polymerization, using ssRNA as a template. In plants, siRNA can induce RNA polymerisation in both 3'®5' and 5'®3' directions, whereas in animals RdRP travels in only one direction (3'®5'), along a certain mRNA, to amplify the silencing signal (Vaistij et al., 2002). Production of dsRNA feeds the initiation phase for the production of more siRNAs, and the process continues in a reiterated fashion. Grafting experiments have proven that systemic dispersion of a silencing signal does occur in plants, being called systemic acquired silencing (SAS). An elegant experiment has shown beta-glucuronidase (GUS) silencing in a GUS-expressing scion grafted onto a GUS-silenced rootstock. The signal moved unidirectionally from source to sink organs, getting across up to a 30-cm wild-type stem grafted between the GUS-expressing scion and the GUS-silenced rootstock (Palauqui et al., 1997). Although systemic dispersion has been proven to occur, the nature of this mobile silencing signal remains elusive. However, based on its specificity, it probably includes a nucleic acid. Imperfect complementarity between target transcripts and small RNAs can also trigger gene silencing. These ~22-nt small RNAs, with imperfect base pairing, belong to a class called micro-RNAs (miRNAs) (previously termed small temporal RNAs, stRNAs). They are also produced by Dicer in the initiation step, and are thought to be involved in translation repression (Jones, 2002) and in controlling developmental pathways in plants (Baulcombe, 2002; Reinhart et al., 2002; Cerutti, 2003; Hunter and Poethig, 2003), acting in specific mRNA destruction by RISC (Tang et al., 2003). The differences between miRNA and siRNA were emphasised by Bartel and Bartel (2003), who generally termed all the small-sized RNAs found in the cell as tiny RNAs. In general terms, miRNAs are transcribed directly from the genome by their own encoding genes, they show high nucleotide identity within related organisms, and they are used by the cell to regulate various genes. On the other hand, siRNAs are originated from viruses, mRNAs, transposons or heterochromatic DNA, with higher nucleotide divergence in related species, and they are used to silence genes nearly identical to those from which they originated. An important question still to be addressed is whether the same type of RISC can be indistinctly programmed by miRNAs and siRNAs, or not. PTGS in plants is not solely triggered by dsRNA directly formed from expressed hpRNA, asRNA, or viruses; it is also elicited by high expression of sense transcripts from a transgene. This phenomenon is called co-suppression, and it is thought to be correlated with the accumulation of free 3' hydroxyl termini that are not covered by poly(A)-binding proteins, leading to the formation of hairpin structures that would instigate the silencing process by the aforementioned mechanisms (Hammond et al., 2001; Lipardi et al., 2001). GENES INVOLVED IN THE PLANT GENE SILENCING MECHANISM Mutations in genes involved in the gene-silencing pathway can be found in individuals who fail to have their genes silenced by PTGS. Interestingly, these mutants are frequently associated with developmental aberrations. Although we still do not fully comprehend the PTGS genetic network, it is clear that PTGS and VIGS pathways share common players, which may also be important elements in normal plant development in the recently disclosed RNA metabolism. We present here an updated set of genes that are involved in gene silencing mechanisms (Table 1), the main features of their gene products, and the phenotypes produced by their mutant alleles in plants.
The gene coding for the enzyme Dicer-1 (DCR1) from Drosophila, which is active in the dsRNA cleavage process, is related to the gene coding RNase III from E. coli. In Arabidopsis, three presumed genes related to DCR1 (SHORT INTEGUMENTS1, SIN1; SUSPENSOR1, SUS1, previously named EMBRYO DEFECTIVE76, EMB76, and CARPEL FACTORY, CAF) were found to be mutant alleles with different strengths of the same gene, and they were renamed DICER-LIKE1 (DCL1; Golden et al., 2002; Schauer et al., 2002). All these DCL1 mutants showed developmental abnormalities. CAF loss-of-function causes overproliferation of the floral meristem (Jacobsen et al., 1999), while the SUS1 and SIN1 mutations are associated, respectively, with embryo abnormalities (Schwartz et al., 1994) and ovule defects (Robinson-Beers et al., 1992). Dicer homologues contain a PAZ domain, which is thought to mediate interactions in the RISC. PAZ is an acronym for the proteins PIWI, ARGONAUTE and ZWILLE (Cerutti et al., 2000). Dicer homologues also bear RNase III motifs and an RNA helicase domain (Hammond et al., 2001). Four Dicer homologues have been found in the Arabidopsis genome, and the allelic series of DCL1 already encompasses 10 mutant members (Golden et al., 2002; Finnegan et al., 2003). Two Dicer homologues have been identified in the rice genome, corroborating with the assumption that the silencing mechanism is also conserved in monocots (Golden et al., 2002). HUA ENHANCER1 (HEN1) was recently discovered in Arabidopsis as a new player in RNA metabolism, being required, together with the Dicer homologue DCL1, for the accumulation of miRNAs (Park et al., 2002). HEN1 is associated with stem, leaf and floral organ growth, with male and female fertility, with floral meristem identity, and with floral determinacy (Chen et al, 2002). An 18-21-nt RNA transcript (called MIR172) was identified in inflorescences, leaves and stems of wild-type Arabidopsis, and it showed sequence complementarity to APETALA2 (AP2), whereas it was barely detected in hen1 and dcl mutants (Park et al., 2002). There is evidence that miRNA metabolism and PTGS are intrinsically related phenomena, since dcl1 and hen1 Arabidopsis mutants have drastically reduced miRNA levels and similar developmental aberrations (Park et al., 2002). A gene called SILENCING DEFECTIVE1 and SUPPRESSOR OF GENE SILENCING2 (SDE1/SGS2) (Dalmay et al., 2000; Mourrain et al., 2000) was identified in PTGS-defective Arabidopsis lines; it codes for an RdRP. In plants, RdRP was first characterized in the tomato (Schiebel et al., 1998). This protein is proposed to synthesize a dsRNA PTGS initiator. When instigated to silence transgenes, sde1/sgs2 mutants produce low levels of correlated tiny RNAs, indicating that this gene is important for transgene PTGS (Dalmay et al., 2000). A virus-induced PTGS was enhanced in these mutants instead, since viruses provide their own RdRP gene in order to produce dsRNA; the cell can recognize this atypical arrangement and trigger its gene silencing mechanism. This result is a strong indication that PTGS is indeed an antiviral defense mechanism that can trigger degradation of RNA derived from exogenous DNA (Mourrain et al., 2000). Together with SGS2, SGS1 has been identified in Arabidopsis mutants that fail to promote PTGS (Elmayan et al., 1998), indicating that the products from both genes are trans-acting elements involved specifically in PTGS. SGS3 is another gene related to the PTGS in Arabidopsis. Its lack of function gives the same phenotype reported above for SGS2 (Mourrain et al., 2000). However, in contrast to SGS2, the SGS3 product is not related to any functionally characterized protein. Recently, SGS3 was found to share sequence homology to the rice gene X, with two conserved domains: an XS domain that indicates a putative enzymatic activity conjugated to coiled-coil structure, suggesting an oligomerization site, and a zf-XS domain, with a still unknown function (Bateman, 2002). SDE3 was found in Arabidopsis as a gene encoding an RNA helicase required for the PTGS pathway (Dalmay et al., 2001). The sde3 mutant showed the same phenotype described for sde1/sgs2, but its function in this proposed mechanism requires further analysis. Likewise, the SDE2 loss-of-function mutant also has a similar phenotype, and its function is still uncharacterized. However, SDE4 has a distinct phenotype, and it may be associated with a step required for DNA methylation (Hamilton et al., 2002). The ARGONAUTE1 (AGO1) gene was identified in Arabidopsis mutants producing altered leaf morphology, a reduced number of secondary meristems, and sterility due to flower developmental defects (Bohmert et al., 1998). AGO1 belongs to a gene family present in many kingdoms, including at least eight members in the Arabidopsis genome (Fagard et al., 2000; Hammond et al., 2001). Two main domains are found in AGO1: the PAZ domain, which may be implicated in protein-protein interactions, and the PIWI domain, which has an unidentified function. Both domains are referred to as the PPD domain. Mutations in other genes of this family induce similar ontogenic defects and PTGS impairment, as is observed in ago1 mutants. Members of the AGO gene family have also been linked to histone modifications and transcriptional gene silencing in several eukaryote species (Hall et al., 2002; Mochizuki et al., 2002; Pal-Bhadra et al., 2002; Taverna et al., 2002; Volpe et al., 2002). In plants, mutation of AGO4 leads to suppression of silencing of SUPERMAN (SUP), a zinc-finger homeotic transcription factor, through its clark kent (clk) epigenetic alleles, rescuing the wild-type phenotype in Arabidopsis. AGO4 appears to be mechanistically related to SDE4, and it is plausible that both are involved in generating long siRNAs, specialized in chromatin level gene silencing (Zilberman et al., 2003), rather than being implicated in VIGS. It is now clear that PTGS is widespread in the diverse kingdoms, sharing conserved factors in species such as Arabidopsis and Drosophila. The challenge now lies in understanding the genetic regulation of gene silencing pathways in order to establish their network interactions. VECTORS FOR VIRUS-INDUCED GENE SILENCING The regions of the plant that can be affected by gene silencing depend mostly upon viral vector characteristics. Silencing vectors usually travel systemically through the phloem in the vascular tissues to most parts of the plant. Although most of the viruses do not penetrate meristems, some have been found to deliver a silencing signal to meristematic regions of the plant (Peele et al., 2001). Some important characteristics of a viral silencing vector are its effectiveness in inducing silencing; its capability to infect and induce silencing in growing parts of the plant; its genome size (due to cloning steps for fragment insertion, since usually the smaller the vector the easier the cloning); its genome partition; the type of nucleic acid its genome is composed of (given that RNA plant viruses are the most common, though DNA viruses can facilitate inoculation as they allow the use of DNA instead of unstable RNA); its host range; whether its genome is or can be made available in binary vectors (to facilitate inoculation via Agrobacterium), and its safety for individuals and the environment. Tobacco mosaic virus (TMV) was the first viral vector used to successfully elicit VIGS of an endogenous gene in a plant species (Kumagai et al., 1995). This work is known as a breakthrough in the plant PTGS research field. Potato virus X (PVX) (Ruiz et al., 1998) followed as the next viral vector used to carry a gene into a plant cell and trigger silencing via VIGS. Although effective, its incapability of infecting meristems is a great disadvantage, since delivering silencing to meristematic tissues is a sine qua non for studying genetic functions involved in developmental pathways. The advent of the Tobacco rattle virus (TRV) vector for VIGS (Ratcliff et al., 2001) opened the possibility of studying genes expressed in early organ development, because of its ability to reach growing points and to deliver the silencing signal to meristems. The cloning of its genome into binary Agrobacterium tumefaciens plasmids immensely facilitated the infection process. TRV is an ssRNA virus with a bipartite genome. The component called RNA1 encodes, among other genes, an RdRP, whereas its genome partner, called RNA2, encodes the coat protein (Angenent et al., 1986; MacFarlane, 1999). The target gene fragment for silencing is inserted into the RNA2 element. Inoculation, either mechanical or via agroinfiltration, requires the presence of both genome components. In the case of agroinfiltration, two different Agrobacterium clones, one carrying the RNA1 genome and another with the RNA2 containing the target gene fragment, are mixed together and co-infiltrated into the leaf tissues (English et al., 1997). Notwithstanding the advantages, such as facility of application with leaf agroinfiltration, prompt results within days after inoculation, and non-dependence on the whole coding sequence of the target gene in order to elicit silencing, for attaining high throughput, there are additional requirements to be met, including a high cloning efficiency, in order to enable a high number of candidate sequences to be placed into the vector within a short period. This condition is fulfilled by a TRV vector based on the Gateway cloning technology (Invitrogen Life Technologies). No restriction enzymes are involved in the Gateway cloning steps; instead, recombination sites that flank the gene fragment are the targets of recombinases, which very efficiently translocate the fragment into the vector (Liu et al., 2002a). A significant limitation of the VIGS system is that TRV, and most of the other vectors available, work very well in N. benthamiana and in the tomato, but they do not show efficacy in Arabidopsis, in which a myriad of genes still need to be functionally analyzed. Additionally, a vector with the capability to infect monocot species, such as maize, rice and lily, is also greatly desired. Figure 2 gives the systematic classification of the more important plant species cited in this text. 
Figure 2. Systematics of species frequently cited in the text for reference. Post-transcriptional gene silencing is well established, especially among the Solanaceae. Efficient virus-induced gene silencing approaches for other organisms, such as Arabidopsis and monocot species are only recently being developed.
The taxonomic information was retrieved (Aug/14/2003) from the Integrated Taxonomic Information System (ITIS) [http://www.itis.usda.gov]. Geminivirus vectors, which have a DNA-based genome, are very promising tools, especially because they enable direct plasmid DNA infection instead of requiring in vitro transcription for mechanical inoculation. The Tomato golden mosaic geminivirus (TGMV) vector (Kjemtrup et al., 1998) effectively silences the PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA), an essential gene associated with DNA replication, in meristematic tissues of N. benthamiana (Peele et al., 2001). Satellite viruses use the genome information contained in helper viruses in order to replicate and move inside the plant. A VIGS system based on the Satellite tobacco mosaic virus (STMV), which uses the TMV strain U2 as a helper, was recently reported (Gosselé et al., 2002). This method is called satellite virus-induced silencing system (SVISS), and it shows some advantages over other systems, such as high stability and accumulation of the silencing signal in the infected tissues, easy cloning due to its small genome size and attenuated symptoms of virus infection. Enzymes involved in several metabolic pathways are effectively silenced, showing clear phenotypic alterations four weeks after inoculation. The Cabbage leaf curl geminivirus (CbLCV) also has great potential as a VIGS vector, since it has been shown to infect and trigger silencing of transgenes and endogenous genes in Arabidopsis. This vector was the first to effectively infect this species, which could mean a major breakthrough for plant genomics (Turnage et al., 2002). The Barley stripe mosaic virus (BSMV) vector successfully induced gene silencing in barley, using phytoene desaturase (PDS) gene fragments derived from barley, rice and maize. In contrast, PDS from N. benthamiana failed to provoke silencing in barley (Holzberg et al., 2002). This result can be attributed to the divergence in nucleotide identity between the PDS genes from barley and N. benthamiana, which is 74%, whereas it is 91 and 89% between barley and the other two monocots that induce gene silencing, rice and maize, respectively. BSMV has a broad host range in the Poaceae family, including rice, maize, wheat, and oats (Brunt et al., 1996), which demonstrates the possibility of using this vector to address functional gene analysis via VIGS in monocot species, particularly interesting for those with high economic value and of great biological relevance as model species. INOCULATION PROCEDURES The first approach designed to deliver the viral genome, harboring a fragment of the gene to be silenced, was through direct cell inoculation with RNA transcripts, since the first vectors were RNA viruses. A vector containing the viral genome, and the gene fragment of interest under the control of a bacterial promoter (for example, SP6, T3 or T7) allows in vitro transcription of large amounts of RNA. Plants are dusted with an abrasive substance (such as carborundum), the RNA solution is applied to the leaves and friction is exerted in order to deliver the RNA into the parenchyma cells. Since geminiviruses are DNA viruses, when they act as VIGS vectors, they have the advantage of eliminating the transcription step, allowing direct inoculation of plasmid DNA. Since in vitro transcription can be economically prohibitive on a large scale, plasmid DNA isolation provides a much cheaper alternative for working with a large number of vectors. Inoculation of plasmid DNA can be performed mechanically, as described above for RNA, or by microbombardment, in which tungsten or gold microprojectiles coated with the plasmid DNA are introduced into seedlings (Kjemtrup et al., 1998; Peele et al., 2001; Turnage et al., 2002). Agroinfection uses Agrobacterium binary vectors to transfer the genetic information into plant cells. In the VIGS system, they are engineered to harbor the viral genome holding the gene of interest. The leaf agroinfiltration method delivers the viral genome into plant cells in order to infect and initiate the transient silencing process by injecting Agrobacterium culture into the leaf parenchyma with a needleless syringe. Despite the efficiency of inoculation and ease of use, leaf agroinfiltration is routine only for a few species, mainly in the Solanaceae family, such as N. benthamiana and the tomato. Additional efforts must be undertaken to determine if this technique can also be applied efficiently in other plant families. In tomato, an alternative approach to leaf agroinfiltration is spraying the bacterial culture on the leaves with a paint airbrush (Liu et al., 2002a). We have induced PDS silencing with a TRV vector cloned in Agrobacterium. While gene silencing with the agroinfiltration method was successful in about 50% of the plants, the airbrush approach resulted in 90 to 100% gene silencing. TARGET GENES AND SILENCING FRAGMENTS IN PTGS AND VIGS The first gene reported to be silenced by PTGS was CHALCONE SYNTHASE (CHS) in petunia, overexpression of which was expected to induce deep purple petals; however, it induced white or variegated flowers instead (Napoli et al., 1990). A similar event occurred with the first gene silencing report in fungi, known as quelling, with the overexpression of the genes involved in carotenoid metabolism causing an albino phenotype, instead of producing the anticipated deep orange color (Cogoni et al., 1994). The PDS gene (Demmig-Adams and Adams, 1992) is ideal to demonstrate the effectiveness of VIGS. This gene participates in the carotenoid metabolic pathway, acting on the antenna complex of the thylakoid membranes, and it protects the chlorophyll from photooxidation. Silencing if this gene results in a drastic decrease in leaf carotene content, leading to a photobleaching symptom that is easily detected. It has been frequently used as a positive control of the system (Ruiz et al., 1998; Ratcliff et al., 2001; Holzberg et al., 2002; Liu et al., 2002a; Turnage et al., 2002). There are several leaf bleaching phenotypes in N. benthamiana that are triggered by PDS fragments from tomato and lily (Figure 3). 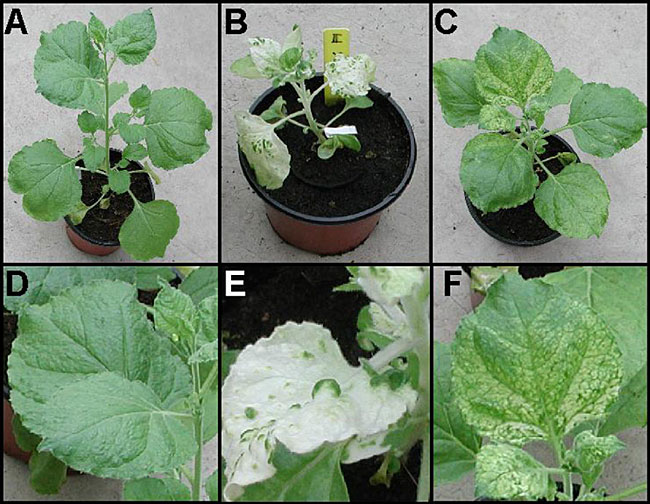
Figure 3. Nicotiana benthamiana at 22 days post-inoculation. Plants were agro-infiltrated with a Tobacco rattle virus-derived vector carrying a phytoene desaturase (PDS) fragment from tomato (B, E) or lily (C, F), using agroinfiltration. Gene silencing can be observed by the leaf bleaching phenotype. Notice the strong silencing triggered by the PDS from tomato, whereas the lily PDS fragment induced much weaker bleaching. Symptoms were already clear at 10 days post-infection. Mock plants are shown in A and D.
Vectors harboring a GREEN FLUORESCENT PROTEIN (GFP) gene fragment have been used occasionally to demonstrate gene silencing in transgenic plants (Ruiz et al., 1998; Peele et al., 2001; Ratcliff et al., 2001; Turnage et al., 2002) and, together with GUS, they are considered the genes of choice for studies on the suppression of gene silencing. Functional characterization of a CELLULOSE SYNTHASE A (CES A) gene series from tobacco (N. tabacum) was undertaken in order to silence their orthologues in N. benthamiana with a VIGS vector based on PVX (Burton et al., 2000). Silencing of this function resulted in a dwarf phenotype, decreased cellulose content in the cell wall, and increased homogalacturan levels. Genes involved in resistance responses against pathogens have also been studied via VIGS. The tobacco Rar1, EDS1 and NPR1/NIM1-like genes were proven to be involved in resistance to TMV by using a TRV-based vector in N. benthamiana (Liu et al., 2002b). In Arabidopsis, CONSTITUTIVE TRIPLE RESPONSE 1 and 2 (CTR1 and 2) encode protein kinases that negatively regulate ethylene responses. Their loss-of-function induces severe dwarfism and constitutive expression of ethylene-inducible genes. VIGS was applied in the tomato to confirm the functional homology between CTR1 and 2 from Arabidopsis and its homologue sequences (Liu et al., 2002a). Silencing of tomato CTR1 and 2 homologues in this species induced the same lack-of-function phenotype observed in Arabidopsis ctr1 mutants. Through co-suppression approaches, it was found that it is not necessary to use the whole coding region in order to induce gene silencing; only a fragment with considerable homology with the target gene is sufficient to elicit the silencing process (Ruiz et al., 1998). Conserved boxes of gene families can potentially trigger silencing of other members concomitantly (Ratcliff et al., 2001). One example is the crossed co-suppression of MADS box transcription factors family in petunia. Overexpression of the PETUNIA FLOWERING GENE (PFG) induces co-suppression of PFG itself and of FLOWERING BINDING PROTEIN 26 (FBP26), which shares high overall sequence homology with PFG (74%), particularly within the MADS box (88%) (Immink et al., 1999). In this way, and in accordance with the molecular model for gene silencing, it is reasonable to postulate that the sequence homologies found in conserved boxes of gene families are more important for multiple gene silencing than are their overall homologies. This may also be true for VIGS. Gene fragments of around 300 to 800 nt have been frequently used in VIGS systems (Ratcliff et al., 2001; Holzberg et al., 2002; Liu et al., 2002a), although smaller fragments (even from 23 to 60 nt), may also be effective if they have considerable homology (Thomas et al., 2001). Gosselé and colleagues (2002), working with the SVISS system, reported that insert sizes larger than 300 nt led to lower levels of gene silencing, whereas fragments of about 100 nt successfully induced silencing. Geminiviruses impose strict limitations on the size of DNA insertions that they can carry. Fragments of 92 to 154 nt of the SULFUR (SU) gene were inserted into the vector, and they were effective in inducing specific gene silencing with TGMV. Additionally, 51 nt of a homologous sequence seemed to be near the lower limit to trigger gene silencing in this system, while gene fragments of 455 and 935 nt were less efficient in triggering gene silencing than were the smaller fragments (Peele et al., 2001). Overexpression of genes transcribing hpRNA of several genes involved in flower developmental pathways leads to specific and heritable silencing of the endogenous genes, resulting in null phenotypes in Arabidopsis (Chuang and Meyerowitz, 2000). VIGS of a regulatory flower developmental gene was first accomplished in N. benthamiana with its LEAFY/FLORICAULA orthologue, NFL (Ratcliff et al., 2001), opening avenues for using this system for efficient characterization of floral gene functions. For optimal results, leading to specific, effective and reliable gene silencing in a heterologous system, the establishment of the level of sequence homology that is required is pivotal information. It is generally assumed that 85% nucleotide identity would be the lowest limit for triggering the silencing mechanism; however, there is no experimental evidence. Although some studies are still necessary before VIGS can be used in the functional identification of MADS-box genes in homo- or heterologous systems, the future seems promising; possibly in the near future this field will be open for speedy and simplified functional characterization of the many genes from new species, for which function has yet to be discovered or confirmed. All these new possibilities and improvements of VIGS are now challenging the system for its efficiency in probing the functions of important genes from various plant species. In our case we mainly study those linked to flower development, such as the MADS-box transcription factors. PERSPECTIVES FOR REVERSE GENETICS OF MONOCOT GENES BASED ON VIGS It has been demonstrated that the gene silencing mechanism is conserved and that the phenomenon can function in monocots (Iyer et al., 2000), even though the VIGS system is not yet well established in these species. The first experiment addressing the induction of transient gene silencing in a monocot species used microprojectile bombardment, a common transformation procedure for monocots, to deliver hpRNA into leaf epidermal cells of maize, wheat and barley (Schweizer et al., 2000). However, researchers already found gene silencing in transgenic plants of several Graminae species a long time ago, including rice, barley, maize, wheat, and sugarcane, besides bulbous monocot species, such as lily, orchids and others (reviewed by Iyer et al., 2000). PTGS was first proven to occur in rice protoplasts expressing the beta-glucuronidase (GUS) gene (Kanno et al., 2000). Moreover, virus resistance in monocot species has been accredited to PTGS in some cases, such as in rice (Pinto et al., 1999), sugarcane (Ingelbrecht et al., 1999) and ryegrass (Xu et al., 2001). Our preliminary results indicate that a PDS fragment derived from a monocot, such as lily, can elicit VIGS in N. benthamiana, in spite of the remote evolutionary relationship between these species (Figure 3). VIGS was induced by a TRV vector, containing a 300-bp PDS fragment, derived from lily. This lily fragment shows 70% identity to the PDS from N. benthamiana. Although the symptoms were not as strong as the bleaching elicited by the tomato PDS fragment, it shows that VIGS can work in a heterologous dicot system using a monocot gene fragment. Despite the convincing results obtained with BSMV in barley (Holzberg et al., 2002), it would be premature to establish this species as a model crop for VIGS in monocots. Rice would be the first option, given its importance as staple food worldwide and as an already established model organism in molecular biology; furthermore, its genome has been completely sequenced. In fact, a novel VIGS vector is about to be released for rice (Ding and Nelson, 2003). The system is based on the Brome mosaic virus, isolated from the grass Festuca pratensis (F-BMV). It is a tetrapartite virus that also infects barley, rice (both japonica and indica) and maize efficiently, and it was engineered to boost gene silencing in grasses. Additionally, this virus has no known insect vector, nor is it transmitted through seeds, increasing the safety of this system. Full reports are still awaited, but preliminary information already shows the potential of this vector for assessing functions from monocot genes, using closely related species as a heterologous system, instead of making use of more divergent, dicot species. These recent reports are providing good prospects for the possibilities of using VIGS for reliable and straightforward reverse genetics in various plant species, including monocots. ACKNOWLEDGMENTS The authors are greatly thankful to Koen Pelgrom, Martin Verbeek and Suzan Gabriels for significant contributions during the course of this work. Anne-Marie Wolters is especially acknowledged for critical reading of the manuscript. Research supported by CNPq, an Organ for Science and Technology Development of the Brazilian Government (200851/98-5), and by the Dutch Ministry of Agriculture, Nature Management and Fisheries (DWK 364). REFERENCES Al-Kaff, N.S., Covey, S.N., Kreike, M.M., Page, A.M., Pinder, R. and Dale, P.J. (1998). Transcriptional and post-transcriptional plant gene silencing in response to a pathogen. Science 279: 2113-2115. Angenent, G.C., Linthorst, H.J.M., van Belkum, A.F., Cornelissen, B.J.C. and Bol, J.F. (1986). RNA2 of tobacco rattle virus strain TCM encodes an unexpected gene. Nucleic Acids Res. 14: 4673-4682. Bartel, B. and Bartel, D.P. (2003). MicroRNAs: at the root of plant development? Plant Physiol. 132: 709-717. Bateman, A. (2002). The SGS3 protein involved in PTGS finds a family. BMC Bioinformatics 3: 21-24 [http://www.biomedcentral.com/1471-2105/3/21]. Accessed August 15, 2002. Baulcombe, D.C. (1999). Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 2: 109-113. Baulcombe, D.C. (2002). An RNA microcosm. Science 297: 2002-2003. Bernstein, E., Caudy, A.A., Hammond, S.M. and Hannon, C.J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M. and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17: 170-180. Brunt, A.A., Crabtree, K., Dallwitz, M.J., Gibbs, A.J., Watson, L. and Zurcher, E.J. (Eds.) (1996). Plant viruses online: descriptions and lists from the VIDE Database [http://biology.anu.edu.au/Groups/MES/vide]. Accessed June 29, 2003. Burton, R.A., Gibeaut, D.M., Bacic, A., Findlay, K., Roberts, K., Hamilton, A., Baulcombe, D.C. and Fincher, G.B. (2000). Virus-induced silencing of a plant cellulose synthase gene. Plant Cell 12: 691-705. Cerutti, H. (2003). RNA interference: travelling in the cell and gaining functions? Trends Genet. 19: 39-46. Cerutti, L., Mian, N. and Baterman, A. (2000). Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 25: 481-482. Chen, X., Liu, J., Cheng, Y. and Jia, D. (2002). HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129: 1085-1094. Chuang, C.-F. and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97: 4985-4990. Cogoni, C. and Macino, G. (2000). Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 10: 638-643. Cogoni, C., Romano, N. and Macino, G. (1994). Suppression of gene expression by homologous transgenes. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 65: 205-209. Covey, S.N., Al-Kaff, N.S., Langara, A. and Turner, D.S. (1997). Plants combat infection by gene silencing. Nature 385: 781-782. Dalmay, T., Hamilton, A., Rudd, S., Angell, S. and Baulcombe, D.C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for post-transcriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543-553. Dalmay, T., Horsefield, R., Braunstein, T.H. and Baulcombe, D.C. (2001). SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20: 2069-2077. Demmig-Adams, B. and Adams, W.W. (1992). Photoprotection and other responses of plants to high light stress. Annu. Rev. Plant. Physiol. 43: 599-626. Ding, X. and Nelson, R. (2003). Analysis of gene function in rice by virus-induced RNA silencing. In: Proceedings of the 7th International Congress of Plant Molecular Biology, June 23-28, 2003, Barcelona, 467: W04-11. Elmayan, T., Balzergue, S., Béon, F., Bourdon, V., Daubremet, J., Guénet, Y., Mourrain, P., Palauqui, J.-C., Vernhettes, S., Vialle, T., Wostrikoff, K. and Vaucheret, H. (1998). Arabidopsis mutants impaired in cosuppression. Plant Cell 10: 1747-1757. English, J.J., Davenport, G.F., Elmayan, T., Vaucheret, H. and Baulcombe, D.C. (1997). Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J. 12: 597-603. Fagard, M., Boutet, S., Morel, J.B., Bellini, C. and Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97: 11650-11654. Finnegan, E.J., Margis, R. and Waterhouse, P.M. (2003). Post-transcriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr. Biol. 13: 236-240. Fire, A., Xu, S.Q., Montgomery, M.K., Kostas, S.A., Driver, S.E. and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806-811. Golden, T.A., Schauer, S.E., Lang, J.D., Pien, S., Mushegian, A.R., Grossniklaus, U., Meinke, D.W. and Ray, A. (2002). SHORT INTEGUMENTS1/ SUSPENSOR1/ CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 130: 808-822. Gosselé, V., Fache, I., Meulewaeter, F., Cornelissen, M. and Metzlaff, M. (2002). SVISS - a novel transient gene silencing system for gene function discovery and validation in tobacco plants. Plant J. 32: 859-866. Guo, S. and Kemphues, K. (1995). par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81: 611-620. Gura, T. (2000). A silence that speaks volumes. Nature 404: 804-808. Hall, I.M., Shankaranarayana, G.D., Noma, K.I., Ayoub, N., Cohen, A. and Grewal, S.I.S. (2002). Establishment and maintenance of a heterochromatin domain. Science 297: 2232-2237. Hamilton, A., Voinnet, O., Chappell, L. and Baulcombe, D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21: 4671-4679. Hamilton, A.J. and Baulcombe, D.C. (1999). A species of small antisense RNA in post-transcriptional gene silencing in plants. Science 286: 950-952. Hammond, S.M., Bernstein, E., Beach, D. and Hannon, C.J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293-296. Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R. and Hannon, C.J. (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146-1150. Holzberg, S., Brosio, P., Gross, C. and Pogue, G. (2002). Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30: 315-327. Hunter, C. and Poethig, R.S. (2003). Missing links: miRNAs and plant development. Curr. Opin. Genet. Dev. 13: 372-378. Immink, R.G.H., Hannapel, D.J., Ferrario, S., Busscher, M., Franken, J., Lookeren Campagne, M.M. and Angenent, G.C. (1999). A petunia MADS box gene involved in the transition from vegetative to reproductive development. Development 126: 5117-5126. Ingelbrecht, I.L., Irvine, J.E. and Mirkov, T.E. (1999). Post-transcriptional gene silencing in transgenic sugarcane. Dissection of homology-dependent virus resistance in a monocot that has a complex polyploid genome. Plant Physiol. 119: 1187-1198. Iyer, L.M., Kumpatla, S.P., Chandrasekharan, M.B. and Hall, T.C. (2000). Transgene silencing in monocots. Plant Mol. Biol. 43: 323-346. Jacobsen, S.E., Running, M.P. and Meyerowitz, E.M. (1999). Disruption of an RNA helicase/RNA III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231-5243. Jones, L. (2002). Revealing micro-RNAs in plants. Trends Plant Sci. 7: 473-475. Kanno, T., Naito, S. and Shimamoto, K. (2000). Post-transcriptional gene silencing in cultured rice cells. Plant Cell Physiol. 41: 321-326. Kjemtrup, S., Sampson, K.S., Peele, C.G., Nguyen, L.V., Conkling, M.A., Thompson, W.F. and Robertson, D. (1998). Gene silencing from plant DNA carried by a geminivirus. Plant J. 14: 91-100. Krysan, P.J., Young, J.C. and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283-2290. Kumagai, M.H., Donson, J., Della-Cioppa, G., Harvey, D., Hanley, K. and Grill, L.K. (1995). Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 92: 1679-1683. Lindbo, J.A., Silva-Rosales, L., Proebsting, W.M. and Dougherty, W.G. (1993). Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5: 1749-1759. Lipardi, C., Wei, Q. and Paterson, B.M. (2001). RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107: 297-307. Liu, Y., Schiff, M. and Dinesh-Kumar, S.P. (2002a). Virus-induced gene silencing in tomato. Plant J. 31: 777-786. Liu, Y., Schiff, M., Marathe, R. and Dinesh-Kumar, S.P. (2002b). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30: 415-429. MacFarlane, S.A. (1999). Molecular biology of the tobraviruses. J. Gen. Virol. 80: 2799-2807. Maeda, I., Kohara, Y., Yamamoto, M. and Sugimoto, A. (2001). Large-scale analysis of gene function in Caenorhabditis elegans by high throughput RNAi. Curr. Biol. 11: 171-176. Mochizuki, K., Fine, N.A., Fujisawa, T. and Gorovsky, M.A. (2002). Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110: 689-699. Mourrain, P., Béclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.B., Jouette, D., Lacombe, A.M., Nikic, S., Picault, N., Rémoué, K., Sanial, M., Vo, T.A. and Vaucheret, H. (2000). Arabidopsis SGS2 and SGS3 genes are required for post-transcriptional gene silencing and natural virus resistance. Cell 101: 533-542. Napoli, C., Lemieux, C. and Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into Petunia results in reversible cosuppression of homologous genes in trans. Plant Cell 2: 279-289. Nishikura, K. (2001). A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell 107: 415-418. Nykänen, A., Haley, B. and Zamore, P.D. (2001). ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309-321. Palauqui, J.-C., Elmayan, T., Pollien, J.-M. and Vaucheret, H. (1997). Systemic acquired silencing: transgene specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16: 4738-4745. Erratum (1998) 17: 2137. Pal-Bhadra, M., Bhadra, U. and Birchler, J.A. (2002). RNAi related mechanisms affect both transcriptional and post-transcriptional transgene silencing in Drosophila. Mol. Cell. 9: 315-327. Parinov, S., Sevugan, M., Ye, D., Yang, W.C., Kumaran, M. and Sundaresan, V. (1999). Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263-2270. Park, W., Li, J., Song, R., Messing, J. and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, an HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484-1495. Peele, C., Jordan, C.V., Muangsan, N., Turnage, M., Egelkrout, E., Eagle, P., Hanley-Bowdoin, L. and Robertson, D. (2001). Silencing of a meristem gene using geminivirus-derived vectors. Plant J. 27: 357-366. Pinto, Y.M., Kok, R.A. and Baulcombe, D.C. (1999). Resistance to rice yellow mottle virus (RYMV) in cultivated African rice varieties containing RYMV transgenes. Nat. Biotechnol. 17: 702-707. Ratcliff, F., Harrison, B.D. and Baulcombe, D.C. (1997). A similarity between viral defense and gene silencing in plants. Science 276: 1558-1560. Ratcliff, F., Martin-Hernandez, A.M. and Baulcombe, D. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25: 237-245. Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B. and Bartel, D.P. (2002). MicroRNAs in plants. Gene Dev. 16: 1616-1626. Robinson-Beers, K., Pruitt, R.E. and Gasser, C.S. (1992). Ovule development in wild-type Arabidopsis and two female sterile mutants. Plant Cell 4: 1237-1249. Rothstein, S.J., Dimaio, J., Strand, M. and Rice, D. (1987). Stable and inheritable inhibition of the expression of nopaline synthase in tobacco expressing antisense RNA. Proc. Natl. Acad. Sci. USA 84: 8439-8443. Ruiz, M.T., Voinnet, O. and Baulcombe, D.C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10: 937-946. Schauer, S.E., Jacobsen, S.E., Meinke, D.W. and Ray, A. (2002). DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 7: 487-491. Schiebel, W., Pelissier, T., Riedel, L., Thalmeir, S., Schiebel, R., Kempe, D., Lottspeich, F., Sanger, H.L. and Wassenegger, M. (1998). Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell 10: 2087-2101. Schwartz, B.W., Yeung, E.C. and Meinke, D.W. (1994). Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 120: 3235-3245. Schweizer, P., Pokorny, J., Schulze-Lefert, P. and Dudler, R. (2000). Double-stranded RNA interferes with gene function at the single-cell level in cereals. Plant J. 24: 895-903. Smith, N.A., Singh, S.P., Wang, M.-B., Stoutjesdijk, P.A., Green, A.G. and Waterhouse, P.M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407: 319-320. Speulman, E., Metz, P.L.J. van, Arkel, G., Hekkert, P.T.L., Stiekema, W.J. and Pereira, A. (1999). A two-component enhancer-inhibitor transposon mutagenesis system for functional analysis of the Arabidopsis genome. Plant Cell 11: 1853-1866. Tang, G., Reinhart, B.J., Bartel, D.P. and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Gene Dev. 17: 49-63. Taverna, S., Coyne, R. and Allis, C. (2002). Methylation of histone H3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110: 701-711. Thomas, C.L., Jones, L., Baulcombe, D.C. and Maule, A.J. (2001). Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J. 25: 417-425. Turnage, M.A., Muangsan, N., Peele, C.G. and Robertson, D. (2002). Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J. 30: 107-114. Vaistij, F.E., Jones, L. and Baulcombe, D.C. (2002). Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA dependent RNA polymerase. Plant Cell 14: 857-867. van der Krol, A.R., Mur, L.A., Beld, M., Mol, J.N.M. and Stuitje, A.R. (1990). Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2: 291-299. Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I.S. and Martienssen, R.A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833-1837. Wang, E. and Wagner, G.J. (2003). Elucidation of the functions of genes central to diterpene metabolism in tobacco trichomes using post-transcriptional gene silencing. Planta 216: 686-691. Waterhouse, P.M., Wang, M.-B. and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411: 834-842. Xu, J., Schubert, J. and Altpeter, F. (2001). Dissection of RNA-mediated ryegrass mosaic virus resistance in fertile transgene perennial ryegrass (Lolium perenne L.). Plant J. 26: 265-274. Zamore, P.D., Tuschl, T., Sharp, P.A. and Bartel, D.P. (2000). RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25-33. Zilberman, D., Cao, X. and Jacobsen, S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716-719. |
|