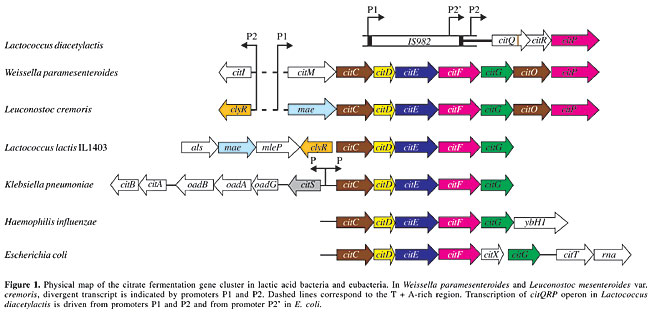
ABSTRACT. Citrate is present in many natural substrates, such as milk, vegetables and fruits, and its metabolism by lactic acid bacteria (LAB) plays an important role in food fermentation. The industrial importance of LAB stems mainly from their ability to convert carbohydrates into lactic acid and, in some species, like Lactococcus lactis and Leuconostoc mesenteroides, to produce C4 flavor compounds (diacetyl, acetoin) through citrate metabolism. Three types of genetic organization and gene locations, involving citrate metabolism, have been found in LAB. Citrate uptake is mediated by a citrate permease, which leads to a membrane potential upon electrogenic exchange of divalent citrate and monovalent lactate. The internal citrate is cleaved into acetate and oxaloacetate by a citrate lyase, and oxaloacetate is decarboxylated into pyruvate by an oxaloacetate decarboxylase, yielding a pH gradient through the consumption of scalar protons. Key words: Lactic acid bacteria, Citrate permease, Citrate lyase, Citrate lyase ligase, Proton motive force, Open reading frame INTRODUCTION Citrate is abundant in nature, and it is a natural constituent of all living cells, being an important source of energy for bacteria. Citrate can be used as a carbon and as an energy source, under both aerobic and anaerobic conditions. Its utilization under aerobic conditions occurs via the tricarboxylic acid cycle, whereas various bacterial fermentation pathways are involved in citrate metabolism under anaerobic conditions (Bott, 1997). A limited number of lactic acid bacteria (LAB) are able to catabolyze carboxylic acids like citrate and malate. Lactic acid bacteria that use citrate play an important role in many dairy processes. In these bacteria, the co-metabolism of citrate and lactose leads to diacetyl and carbon dioxide production. Diacetyl is essential for the flavor of butter, fresh cheese and buttermilk. Carbon dioxide is responsible for cavity formation in certain types of cheese, like blue cheeses. The homofermentative Lactococcus lactis subsp. lactis biovar diacetylactis (L. diacetylactis), and the heterofermentative Leuconostoc species are the main LAB using citrate found in dairy starters. Studies made of citrate metabolism in L. lactis and Leuconostoc mesenteroides have revealed the same genetic pathway, composed of three steps for citrate conversion into pyruvate. The cit genes are positively regulated by pH in Lactococcus and by citrate in Leuconostoc. Energetically, in Leuconostoc the coupling between the citrate metabolic pathway and glycolysis is at the level of the redox state of the cell, and in Lactococcus it is at the level of the end product of glycolysis (Konings, 2002). We reviewed the genetic organization and expression of genes involved in citrate metabolism in LAB. BIOENERGETICS AND MECHANISMS OF CITRATE UPTAKE IN THE LACTIC ACID BACTERIA The mechanism of citrate metabolism in L. lactis and L. mesenteroides is a secondary proton motive force (PMF)-generating pathway. The anionic form of the acid is transported into the cell by an electrogenic secondary carrier generating an exchange with the end-product of the pathway (Marty-Teysset et al., 1995). The PMF results from a pH gradient and the generation of a membrane potential. The trans-membrane pH gradient is due to scalar proton consumption in the decarboxylation of oxaloacetate. The membrane potential-generating secondary transporters, malate permease (MleP) and citrate permease (CitP) involved in malolactic and citrolactic fermentations, translocate net negative charges across the membrane, and they catalyze a heterologous exchange of two structurally related precursor and product molecules, i.e., malate/lactate and citrate/lactate. Lactate is an inside substrate of MleP and CitP and malate could be an outside substrate of CitP (Marty-Teysset et al., 1995). Both MleP and CitP transport a range of 2-hydroxycarboxylates (symbol R1R2COHCOOH) with R substituents ranging in size from two hydrogens (glycolate) to acetyl and methyl groups (citromalate) for MleP and two acetyl groups (citrate) for CitP. The citrate metabolic pathway described in Lactococcus and Leuconostoc is a precursor/product exchange system. The product (lactate) results from the conversion of citrate and also from glucose metabolism, which explains why citrate uptake by Lactococcus and Leuconostoc is efficient only during lactate production from co-metabolism with a sugar. The use of citrate by L. lactis and Leuconostoc spp. strains is strongly dependent on the pH of the medium (Marjo et al., 1991). In Leuconostoc spp. strains, the growth rate is stimulated and CitP expression levels are increased when citrate is present in the medium (Marty-Teysset et al., 1996), whereas growth stimulation by citrate was not observed in L. lactis, while the low constitutive expression of citrate transport increased at low pH (López et al., 1998; Garcia-Quintans et al., 1998). The co-metabolism of citrate and glucose increases the specific rate and molar growth yield of L. mesenteroides and the citrate alkalinizes the external pH (Marty-Teysset et al., 1996). In Leuconostoc, lactate is produced from citrate during citrolactic fermentation, and CitP catalyzes a precursor/product exchange, while in Lactococcus, the pyruvate produced from citrate is not the main precursor of lactate, which is produced via the homofermentative pathway. In Leuconostoc, glucose and citrate fermentation results in a growth advantage relative to growth on glucose alone, because of a metabolic shift in the heterofermentative pathway for glucose. In the absence of citrate, acetyl-phosphate formed from glucose is reduced to ethanol, which balances the redox equivalents from the phosphoketolase pathway. In the presence of citrate, the redox equivalents are shuttled to pyruvate that is produced from citrate, yielding lactate, and acetyl-P is converted into acetate via the acetate kinase pathway, resulting in the generation of one ATP per mole of acetyl-P. GENETIC ORGANIZATION AND EXPRESSION OF CITP IN LACTIC ACID BACTERIA The CitP sequences available in the database are from: Lactococcus lactis subsp. lactis, Leuconostoc lactis, Weissella paramesenteroides, and Leuconostoc mesenteroides subsp. mesenteroides 19D. These CitPs are composed of 442, 441, 442, and 443 amino acids, respectively; they are highly homologous (>98% identity) with a hydrophobic N-terminal domain, and all of them are plasmid-encoded. The loss of the plasmid harboring CitP encoding DNA, by curing experiments, resulted in the interruption of citrate uptake and therefore in diacetyl production, which proves that the citrate transport system is a crucial step. Restoration of this activity is experimentally possible, as observed in a plasmid-free strain L. lactis IL1403 (cit–), which was rendered a diacetyl producer (Bourel et al., 1996), upon its transformation with plasmid pFL3 containing the lactococcal citP gene, under control of the S. pneumoniae polA promoter (Magni et al., 1994). The genetic organization and genomic location of CitP have been established in L. diacetylactis, W. paramesenteroides, and L. mesenteroides subsp. cremoris (L. cremoris) (Figure 1). In lactococcal plasmid pCIT264, citP is included in the citQRP operon, in which citP codes for a CitP, citR codes for a regulatory protein, CitR, and citQ codes for a leader peptide, CitQ (López de Felipe et al., 1995). In L. diacetylactis, transcription of citQRP operon is driven mainly from promoter P1, yielding a 2.9-kb mRNA regulated at the post-transcriptional level by processing that occurs in a complex secondary structure (López de Felipe et al., 1995). Similar processing has been found in the heterologous E. coli (Drider al., 1998), and RNases responsible for this processing were identified by using mutant strains deficient in endoribonucleases (RNase E and RNase III) and exoribonucleases (PNPase and RNase II). The cleavage located at the CitR Shine-Dalgarno sequence is ascribed to RNase III, which seems therefore to play a major role in the control of CitP expression (Drider et al., 1999). In L. lactis, the existence of RNase III was reported for the first time by Drider et al. (1999), and its structural gene was cloned and expressed in E. coli (Drider al., 2002). Moreover, transcription of the citQRP operon is induced by acid stress since the mRNA species detected in the 5’ end region were 14-fold more abundant at pH 4.5 than at pH 6.5 (Garcia-Quintans et al., 1998). At low pH, L. diacetylactis relieves the inhibition due to lactate accumulation by a citrate metabolic pathway induction, making the cells more resistant (Magni et al., 1999). In L. mesenteroides, the existence of the citQRP operon is not established, although the DNA sequence analysis of the upstream and adjacent regions of the citP gene showed a citR disrupted by several mutations (Vaughan et al., 1995). In W. paramesenteroides, the citP gene is included in a plasmidic cluster named citMCDEFGRP, which is transcribed as an 8.8-kb polycistronic mRNA (Martin et al., 1999, 2000). As in L. diacetylactis, the citP gene is immediately preceded by an open reading frame (ORF) encoding a 30-kDa polypeptide termed CitR, whose C-terminal domain is extremely similar (97%) to Lactococcal CitR; but its N-terminal domain is homologous to esterases and peroxidases. Furthermore, two ORFs, citM and citI, encoding for an NAD-dependent malic enzyme and a putative regulatory protein have been identified upstream of citC. In L. cremoris, citP is chromosomally located within a cluster of eight genes named maecitCDEFGOP. Two transcripts of 5.2 kb (citmaeCDEFG) and 4 kb (citGOP), presumably resulting from the processing of a larger transcript, were detected only when cells were grown in the presence of citrate (Bekal et al., 1998; Bekal-Si-Ali et al., 1999). Two ORFs referred to as mae and clyR, whose products are exactly similar to those of citM and citI, were identified upstream of citC. The clyR and citI genes are transcribed divergently from the citmaeCDEFG and citMCDEFG clusters. The putative regulatory proteins ClyR (312 amino acids) and CitI (322 amino acids) belong to the SorC transcriptional regulator family, and the DNA region between the start of citI and citM or clyR and mae contains two putative promoters with an extraordinarily high A + T content (80%). The effect of citI gene on expression of cit operon was studied in E. coli, in which cis or trans addition of the citI gene increased the activity of the cit promoter, showing that transcriptional regulation of citrate utilization is through an activator (Martin et al., 2000). Furthermore, gel shift and footprinting assays revealed that CitI recognizes at least three operator sites placed in the citI-citM intergenic DNA promoter region (Sender et al., 2002). In K. pneumoniae, the transport of citrate is catalyzed by three secondary carriers, called CitS, CitH and CitW. CitS is a Na+-dependent citrate carrier that catalyzes the electroneutral transport of HCit2–, using DpNa and DpH as driving forces (Pos and Dimroth, 1996). CitH is a Na+-independent transporter that catalyzes the Hcit2– in symport with protons (Van der Rest et al., 1991), and it is assumed that CitH is functional under oxic growth conditions. The function of CitW remains to be determined.
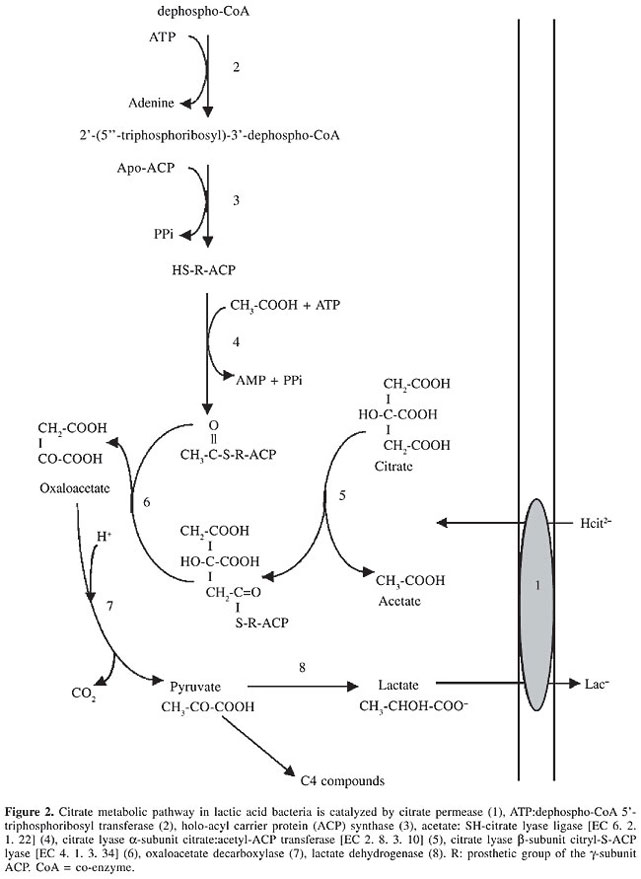
GENETIC ORGANIZATION AND EXPRESSION OF CITRATE LYASE IN LACTIC ACID BACTERIA Different mechanisms of regulation of bacterial citrate lyase (CL), such as configurational changes, reversible covalent modification by acetylation/deacetylation, and phosphorylation/dephosphorylation, have been reported. The CL (EC 4.1.3.6) of L. lactis and Ln. mesenteroides were shown to form a functional complex (Mr 585,000) of three proteins: an acyl carrier protein [ACP] (g-subunit) carrying a prosthetic group; a citrate:acetyl-ACP transferase (a-subunit, EC 2.8.3.10) and a citryl-S-ACP lyase (b-subunit, EC 4.1.3.34) in a stoichiometric relationship of 6:6:6. The structures and the mechanism of action are similar to those of the CL of K. pneumoniae (Subramanian and Sivaraman, 1984; Antranikian and Giffhorn, 1987). The structure of the prosthetic group of CL purified from Klebsiella is a 5-phosphoribosyl-dephospho-co-enzyme A (CoA) attached by its ribose-5-phosphate moiety via a phosphodiester linkage to a serine residue of the ACP. The synthesis and attachment of the prosthetic group involve two reactions. A triphosphoribosyl-dephospho-CoA synthase (ATP:dephospho-CoA 5’-triphosphoribosyl transferase) catalyses via an a-1,2-glycosidic linkage between ATP and dephospho-CoA the formation of the prosthetic group precursor 2’-(5”-triphosphoribosyl)-3’-dephospho-CoA (Figure 2, reaction 2). The second reaction (Figure 2, reaction 3), which consists of the transfer of the prosthetic group precursor to apo-ACP, is catalyzed by a holo-ACP synthase (Schneider et al., 2000a,b). The CL is active only if the thioester residue of the prosthetic group linked to its acyl carrier protein (g-subunit) is acetylated. This activation is catalyzed by an acetate: SH-CL ligase (CLL, EC 6.2.1.22), which converts HS-ACP with ATP and acetate into the acetyl-S-ACP (Figure 2, reaction 4) (Schmellenkamp and Eggerer, 1974). The breakdown of citrate to acetate and oxaloacetate involves two consecutive steps. The a-subunit exchanges the acyl group for a citryl group to form the citryl-S-ACP (Figure 2, reaction 5). Lastly, the b-subunit cleaves citryl-S-ACP into oxaloacetate and regenerates the acyl-S-ACP (Figure 2, reaction 6) (Dimroth and Eggerer, 1975). In LAB, the genes encoding for CLL and CL are part of a cluster designed as citCDEFG. The proteins deduced from these citC, citD, citE, citF are CLL, and the subunits g, b, and a of CL, respectively. CitC catalyzes the ATP-dependent acetylation of the phosphoribosyl dephospho-CoA group of CL. Contrarily to L. lactis, in L. cremoris and W. paramesenteroides, the citMCDEFGRP operon is unequivocally induced by citrate at the transcriptional level, independently of the pH of the medium (Bekal-Si-Ali et al., 1999; Martin et al., 2000). Northern blot analysis revealed an mRNA transcript of 8.8 kb starting upstream of citM and ending downstream of citP; this full-length transcript of 8.8 kb is subjected to a processing taking place in four different secondary structures, leading to synthesis and expression of various proteins at suitable concentrations (Martin et al., 2000). The function of CitG is not yet established in LAB, although sequence alignment showed that its C and N terminal domains are similar to the C-terminal domain of CitX and the N-terminal domain of CitG from K. pneumoniae. In this bacterium, the reactions of the tricarboxylic acid cycle are operative and the presence of citrate synthase requires a strict regulation of CL activity to avoid futile cycling between citrate fermentation and the L-glutamate biosynthetic pathway. After citrate depletion from the growth medium or upon transfer from an anaerobic citrate medium to an aerobic glucose medium, L-glutamate synthesis is ensured from oxaloacetate and acetyl-CoA via citrate only if the citrate fermentation pathway is turned off or partially turned off. The intracellular L-glutamate concentration controls these pathways by modulating the activity of the CL complex (Antranikian and Giffhorn, 1987; Antranikian and Gottschalk, 1989). In K. pneumoniae the citCDEFG cluster is located divergent to citS (Figure 1). Remarkably, in E. coli a citCDEFXG exists and displays high similarity to the CL cluster from K. pneumoniae, but the E. coli CL cluster contains an additional ORF named citX (Schneider et al., 2002). In both bacteria, the citC operon is regulated by a two-component regulatory system called CitA-CitB (Meyer et al., 1997) or DpiB-DpiA (Ingmer et al., 1998). Recently, it was demonstrated that CitG functions as ATP:dephospho-CoA 5’-triphosphoribosyl transferase) and CitX functions as 2’-(5”-triphosphoribosyl)-3’-dephospho-CoA transferase) (Schneider et al., 2002).
CONCLUDING REMARKS AND FUTURE PERSPECTIVES The DNA sequence of L. lactis IL1403 genome (Bolotin et al., 2001) did not reveal any evident ORF matching with oxaloacetate decarboxylase. Conversion of oxaloacetate to pyruvate is likely carried out by the product of mae, since a high level of sequence similarity exists between the mae product and malic enzyme from Bacillus stearothermophilus, an enzyme that catalyzes decarboxylation of oxaloacetate. To confirm this hypothesis, we suggest a study of the impact of the mae product on diacetyl production using a strain deficient in mae ORF. In this mini-review, we have mentioned that CitP of L. cremoris is chromosomally located; this is the first report on such location in LAB. Furthermore, the CitP of L. cremoris appears to be slightly different from those described in the literature. ACKNOWLEDGMENTS The authors are indebted to Prof. Charles Diviès for stimulating work on citrate metabolism in lactic acid bacteria performed in his laboratory during many years. The authors thank Paloma López for her contribution in understanding genetic aspects of citrate metabolism. REFERENCES Antranikian, G. and Giffhorn, F. (1987). Citrate metabolism in anaerobic bacteria. FEMS Microbiol. Rev. 46: 175-198. Antranikian, G. and Gottschalk, X. (1989). Phosphorylation of citrate lyase ligase in Clostridium sphenoides and regulation of anaerobic citrate metabolism in other bacteria. Biochemistry 71: 1029-1037. Bekal, S., Van Beeumen, J., Samyn, B., Garmyn, D., Henini, S., Diviès, C. and Prévost, H. (1998). Purification of Leuconostoc mesenteroides citrate lyase and cloning and characterization of the citCDEFG gene cluster. J. Bacteriol. 180: 647-654. Bekal-Si-Ali, S., Diviès, C. and Prévost, H. (1999). Genetic organization of the citCDEF locus and identification of mae and clyR genes from Leuconostoc mesenteroides. J. Bacteriol. 181: 4411-4416. Bolotin, A., Wincker, P., Mauger, S., Jaillon, O., Malarme, K., Weissenbach, J., Ehrlich, S.D. and Sorokin, A. (2001). The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11: 731-753. Bott, M. (1997). Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 167: 78-88. Bourel, G., Bekal, S., Diviès, C. and Prévost, H. (1996). Citrate permease gene expression in Lactococcus lactis subsp. lactis strains IL 1403 and MG1363. FEMS. Microbiol. Lett. 154: 367-370. Dimroth, P. and Eggerer, H. (1975). Isolation of sub-units of citrate lyase and characterization of their function in the enzyme complex. Proc. Natl. Acad. Sci. USA. 72: 3458-3462. Drider, D., Santos, J.M., Arraiano, C.M. and López, P. (1998). RNA processing is involved in the post-transcriptional control of citQRP operon from Lactococcus lactis biovar diacetylactis. Mol. Gen. Genet. 258: 9-15. Drider, D., Santos, J.M., Garcia-Quintans, N., Arraiano, C.M. and López, P. (1999). The role of Escherichia coli RNase E and RNase III in the processing of the citQRP operon mRNA from Lactococcus lactis biovar diacetylactis. J. Mol. Microbiol. Biotechnol. 2: 337-346. Drider, D., Bolotine, A., Renault, P. and Prévost, H. (2002). Functional study of Lactococcus lactis RNase III in Escherichia coli. Plasmid 47: 246-250. Garcia-Quintans, N., Magni, C., de Mendoza, D. and López, P. (1998). The citrate transport system of Lactococcus lactis subsp. lactis biovar diacetylactis is induced by acid stress. Appl. Environ. Microbiol. 64: 850-857. Ingmer, H., Miller, C.A. and Cohen, S.N. (1998). Destabilized inheritance of pSC101 and other Escherichia coli plasmids by DpiA, a novel two-component system regulator. Mol. Microbiol. 29: 49-59. Konings, W.N. (2002). The cell membrane and the struggle for life of lactic acid bacteria. Anton. Leeuw. Int. J.G. 82: 3-27. López, P., Drider, D., Garcia-Quintans, N., Corrales, M.A., Magni, C., Martin, M. and de Mendoza, D. (1998). Regulation of expression of the Lactococcus lactis subsp. lactis biovar diacetylactis citrate transport system. Dairy Science and Technology (Le Lait) 78: 11-16. López de Felipe, F., Magni, C., de Mendoza, D. and López, P. (1995). Citrate utilization gene cluster of the Lactococcus lactis biovar diacetylactis: organization and regulation of expression. Mol. Gen. Genet. 246: 590-599. Magni, C., López de Felipe, F., Sesma, F., López, P. and de Mendoza, D. (1994). Citrate transport in Lactococcus lactis biovar diacetylactis: expression of the plasmid-borne citrate permease P. FEMS Microbiol. Lett. 118: 75-82. Magni, C., de Mendoza, D., Konings, W.N. and Lolkema, J.S. (1999). Mechanism of citrate metabolism in Lactococcus lactis: Resistance against lactate toxicity at low pH. J. Bacteriol. 181: 1451-1457. Marjo, J., Starrenburg, C. and Hugenholtz, J. (1991). Citrate fermentation by Lactococcus and Leuconostoc spp. Appl. Environ. Microbiol. 57: 3535-3540. Martin, M., Corrales, M.A., de Mendoza, D., López, P. and Magni, C. (1999). Cloning and molecular characterization of the citrate utilization citMCDEFGRP cluster of Leuconostoc paramesenteroides. FEMS Microbiol. Lett. 174: 231-238. Martin, M., Magni, C., López, P. and de Mendoza, D. (2000). Transcriptional control of the citrate-inducible citMCDEFGRP operon encoding genes involved in citrate fermentation in Leuconostoc paramesenteroides. J. Bacteriol. 182: 3904-3912. Marty-Teysset, C., Lolkema, J.S., Schmitt, P., Diviès, C. and Konings, W.N. (1995). Membrane potential-generating transport of citrate and malate catalyzed by CitP of Leuconostoc mesenteroides. J. Biol. Chem. 270: 25370-25376. Marty-Teysset, C., Lolkema, J.S., Schmitt, P., Diviès, C. and Konings, W.N. (1996). The citrate metabolic pathway in Leuconostoc mesenteroides: expression, amino acid synthesis, and a-ketocarboxylate transport. J. Bacteriol. 178: 6209-6215. Meyer, M., Dimroth, P. and Bott, M. (1997). In vitro binding of the response regulator CitB and of its carboxy-terminal domain to A + T rich DNA target sequences in the control region of the divergent citC and citS operons of Klebsiella pneumoniae. J. Mol. Biol. 269: 719-731. Pos, K.M. and Dimroth, P. (1996). Functional properties of the purified Na+-dependent citrate carrier of Klebsiella pneumoniae: evidence for asymmetric orientation of the carrier protein in proteoliposomes. Biochemistry 35: 1018-1026. Schmellenkamp, H. and Eggerer, H. (1974). Mechanism of enzymic acetylation of des-acetyl citrate lyase. Proc. Natl. Acad. Sci. USA 71: 1987-1991. Schneider, K., Dimroth, P. and Bott, M. (2000a). Biosynthesis of the prosthetic group of citrate lyase. Biochemistry 39: 9438-9450. Schneider, K., Dimroth, P. and Bott, M. (2000b). Identification of triphospho-ribosyl-dephospho-CoA as precursor of the citrate lyase prosthetic group. FEBS Lett. 483: 165-168. Schneider, K., Kästner, C.N., Meyer, M., Wessel, M., Dimroth, P. and Bott, M. (2002). Identification of a gene cluster in Klebsiella pneumoniae which includes citX, a gene required for biosynthesis of the citrate lyase prosthetic group. J. Bacteriol. 184: 2439-2446. Sender, P., Martin, M., Lopez, P., de Mendoza, D. and Magni, C. (2002). Characterization of the Weissella citrate operon promoter contribution of CitI and its novel helix-turn-helix motif to promoter activity. Abstract of the Seventh Symposium on Lactic Acid Bacteria, Egmond aan Zee, The Netherlands, FEMS ed. (2002). Subramanian, C. and Sivaraman, C. (1984). Bacterial citrate lyase. J. Biosci. 6: 379-401. Van der Rest, M.E., Abee, T., Molenaar, D. and Konings, W.N. (1991). Mechanism and energetics of citrate-transport system of Klebsiella pneumoniae. Eur. J. Biochem. 195: 71-77. Vaughan, E.E., David, S., Harrington, A., Daly, C., Fitzgerald, G.F. and de Vos, W.N. (1995). Characterization of plasmid-encoded citrate permease (citP) genes from Leuconostoc species reveals high sequence conservation with the Lactococcus lactis citP gene. Appl. Environ. Microbiol. 61: 3172-3176. |
|