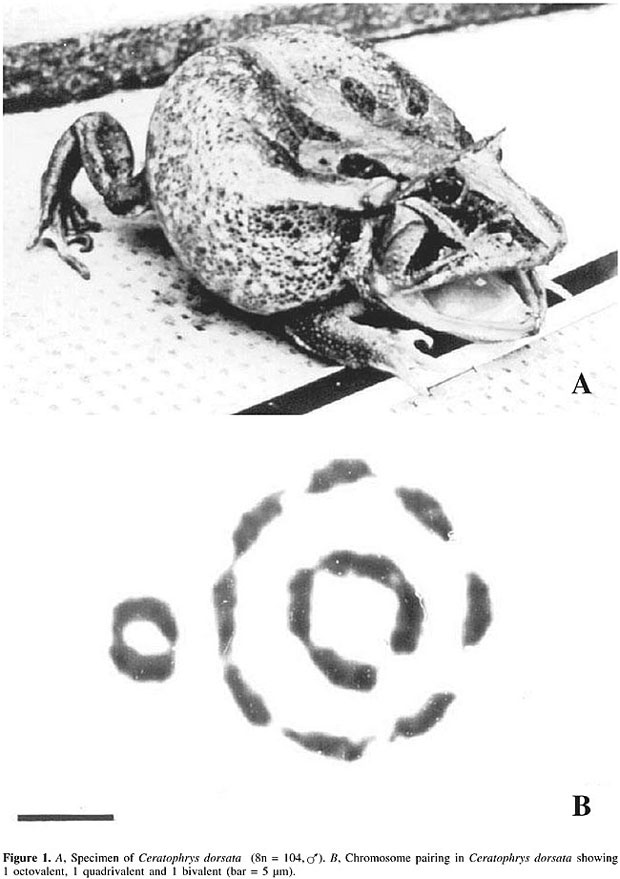
ABSTRACT. The evolution of the metazoa has been characterized by gene redundancy, generated by polyploidy, tandem duplication and retrotransposition. Polyploidy can be detected by looking for duplicated chromosomes or segments of orthologous chromosomes in post-polyploid animals. It has been proposed that the evolutionary role of polyploidy is to provide extra-copies of genes, whose subsequent alteration leads to new functions, increased biological complexity, and, ultimately, speciation. We review the theory of evolution by genome duplication, basing our arguments on findings from autopolyploid anurans and fish, undergoing post-polyploidy diploidization. We conclude that: 1) the high genetic variability of autotetraploid anurans is a result of tetrasomic expression, based on studies of isozymes and other proteins. 2) Epigenetic mechanisms mediate the reduced expression or silencing of redundant copies of genes in the regulation of gene expression of these tetraploids. This conclusion is based on data concerning ribosomal and hemoglobin gene activity. 3) Duplication of the genome may have occurred more than once in the phylogeny of the anurans, as exemplified by 4n and 8n Leptodactylidae species. Key words: Amphibians, Evolution, Gene redundancy, Polyploidy, Speciation INTRODUCTION ‘In a strict sense, nothing in evolution is created de novo’ Ohno (1970). Animal evolution is characterized by an increase in organic complexity generated by the acquisition of new proteins with specific functions. A central problem in genetic evolution is the elucidation of the mechanisms that modulate genomes by the creation, interaction and fixation of the novel proteins that lead to diversification and speciation. Neo-Darwinism assumes that a new species emerges by the gradual accumulation of small mutations and the elimination of those that are deleterious or lethal, by selection. The rapid emergence of a new species is well known in plants, where it occurs by the duplication of hybrid genomes (allopolyploidy). In some animal groups it is difficult to explain the rapid diversification of species and the organic innovations that take place solely through micro-evolution, such changes being more easily explainable by the concept of rapid evolution. The theory of evolution by polyploidy proposed by Ohno (1970) assumes that the vertebrates evolved through mechanisms that generate gene redundancy. The abrupt duplication of a genome would be instrumental in increasing the genetic material, alterations to which, when subjected to selection, could lead to new functions, increased biological complexity and the diversification of species, with increased ecological conquests. Various studies, using techniques from molecular biology, are being used to clarify when the genomic duplications occurred and if they resulted in the acquisition of new functions and increased biological complexity. Within this scenario it has been proposed that genomic duplication has an important role in species diversification, as has been suggested by the divergent resolution model (Lynch and Conery, 2000). The evolutive polyploidy hypothesis is supported by genetic data, which show that the number of genes coded for in vertebrates is four times that found in invertebrates such as Caenorhabditis elegans, Ciona intestinalis and Drosophila melanogaster (Simmen et al., 1998), and also by the ‘tetralogy’ theory of Ohno (1970, 1999) who has shown that various genes of invertebrates such as Drosophila have only one allele, while in vertebrates such as humans there are three or four alleles (Spring, 1997). The alternative hypothesis that the increased gene redundancy of vertebrates is due to duplication of regions of their chromosomes is based on the phylogenetic analysis of 35 gene families made by Martin (1999). Thirty years ago the study of evolutive cytogenetics showed the first example of saltatorial evolution in vertebrates with the discovery of autopolyploidy in bisexual anurans of the families Leptodactylidae and Hylidae. The species of these families have various ploidy levels, 2n, 4n, and 8n (Beçak et al., 1966, 1967, 1970a; Bogart, 1967). The observation that some fish species also show signs of ancestral polyploidy, that is post-polyploidy, detected by the presence of some residual multivalents (Ohno et al., 1968; Wolf et al., 1969), but with high DNA content values (Ohno and Atkin, 1966; Atkin and Ohno, 1967) suggested the evolutive polyploidy hypothesis formulated in 1970 by Ohno. According to Ohno’s hypothesis, two genomic duplications occurred in the evolution of the vertebrates: one in the transition of the invertebrates to the vertebrates and the other during the Devonian diversification of fish. Genome duplication would have ceased after the development of reptiles because of the appearance of morphologically distinct sex chromosomes. Later, however, molecular data on Hox genes lead Ohno (1996, 1999) to accept that the second duplication occurred before the Devonian; this conclusion, reinforced by the paleontological discovery of Cephalochordata in the Cambrian, led Ohno (1996, 1999) to suggest the Cambrian Pananimalia genome hypothesis, which postulates that for the majority of phyla all animals emerged during the Cambrian explosion, which lasted 10 million years. We summarize the findings on autopolyploidy in anurans that lead to the theory of evolution by genome duplication and the mechanisms of gene regulation in autotetraploid anurans. Analysis of these topics suggests that the high genetic variability of autotetraploid anurans is a result of tetrasomic expression and that redundant copies of genes in autotetraploid anurans can be silenced, or have their expression reduced, and some mechanisms of gene repression are epigenetic. POLYPLOIDY AS A FACTOR IN ANURAN EVOLUTION ‘The history of the earth is recorded in the layers of its crust; the history of all organisms is inscribed in the chromosomes’ (Hitoshi Kihara, 1920, cited by Naruya, 2002). Polyploidy, in the form of allopolyploidy, resulting from the duplication of the genome in interspecific hybrids, is of great importance in the diversification of higher plant species, because it results not only in an increase in the amount of DNA and of gene regulation mechanisms, but also in modifications in regulatory interactions as well as genetic and epigenetic changes (Osborn et al., 2003; Adams et al., 2003). From a cytological point of view, allopolyploidy succeeded in plants due to mechanisms that control the segregation of related homologous chromosomes during meiosis. It is known that in plants and amphibians prophase I meiotic nuclei have the cytological appearance of a ‘bouquet’, in which the telomeres agglomerate at the periphery of the nucleus. In allopolyploid plants, this agglomeration is preceded by the pairing of homologue chromosome centromeres. Then the synaptonemal complexes form from the telomeres. It has also been shown that the correct segregation of homologous chromosomes is genetically controlled and dependent on the Ph1 locus; this could be one or more genes or areas of heterochromatin with epigenetic effects (Moore, 2002). Allopolyploidy is also known in parthenogenetic vertebrates, but in the case of bisexual animals it has been suggested that autopolyploidy did not have a relevant role in evolution because chromosomal duplication would be incompatible with the sex-determination mechanisms (Muller, 1925). However, this concept has been superseded with the demonstration of evolution through polyploidy in bisexual anurans (Beçak et al., 1966, 1967, 1970a; Bogart, 1967). The structure of the anuran karyotype has also evolved by chromosome rearrangements, such as centric fusion (Beçak, 1968) and post-polyploid translocations (Beçak and Beçak, 1974b, 1998). Currently, 19 polyploid amphibians are known (Kawamura, 1984). Autopolyploidy has been detected in the anuran families Leptodactylidae and Hylidae by the direct observation of duplicated mitotic chromosomes and multivalent meiotic chromosomes in tetraploid and octoploid animals (Figures 1 and 2); these cytological findings were confirmed by cytophotometric estimates of DNA content (Beçak et al., 1967, 1970). Variation in chromosome number in the genus Odontophrynus (Salientia, Ceratophrydidae) has been interpreted as being due to multiple translocations (Saez and Brum-Zorilla, 1966) and it has also been found that these polyploids are similar to 2n cryptic species (Beçak et al., 1970b; Batistic et al., 1975). Artificial triploid (3n = 33) hybrids have been obtained with Odontophrynus by the mating of tetraploid Odontophrynus americanus females with diploid (2n = 22) O. americanus, O. cultripes and O. carvalhoi males (Beçak et al., 1968a; Beçak and Beçak, 1974a). The segregation of trivalents during meiosis of 3n produces n, 2n and 3n gametes and aneuploids, indicating that triploids may be a stepping-stone for the production of stable 4n tetraploids (Beçak and Beçak, 1970). Other diplo-tetraploid anuran species have also been reported (Bogart and Wasserman, 1972; Bachmann and Bogart, 1975).
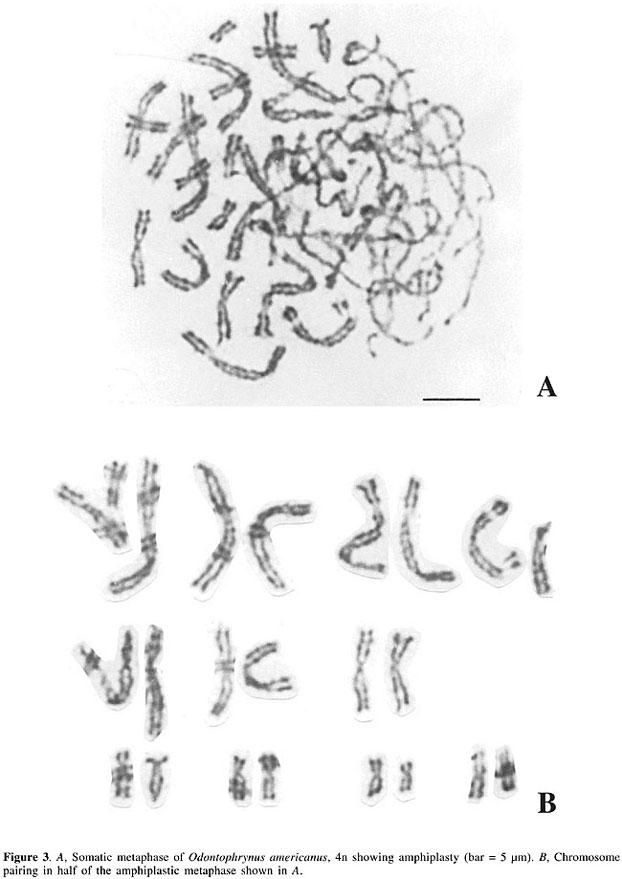
The cytological observation of meiotic tetravalents suggests that chromosome duplication is a relatively recent event, given that diploid and tetraploid O. americanus specimens are morphologically similar (Beçak et al., 1966). Certain intra- and interspecific cytogenetic polymorphisms detected in secondary constrictions (Beçak and Beçak, 1974a) or in banding-patterns of the nucleolus organizer region (NOR) or C-banding regions in O. americanus (Ruiz et al., 1980, 1981; Schmid et al., 1985; Almeida et al., 1986), and in Ceratophrys ornata (Schmid et al., 1985), as well as in Ceratophrys dorsata (Soares-Scott et al., 1988), have been interpreted by Schmid et al. (1985) as post-polyploid alterations. Phylogenetic trees have been proposed for Odontophrynus based on polymorphisms of secondary constrictions (Beçak and Beçak, 1974a) and NOR and C-banding patterns (Ruiz and Beçak, 1976; Ruiz et al., 1980, 1981). The molecular organization of ribosomal cistrons analyzed in population studies on Odontophrynus has also shown polymorphisms from 2n to 4n (Cortadas and Ruiz, 1988). Comparative studies using optical and electron microscopy carried out to examine meiosis and chromosome ultrastructure in diploid and tetraploid amphibians have shown that in tetraploid species the four homologous meiotic chromosomes pair two by two, starting at the extremities of the chromosomes arranged in a ‘bouquet’ (Beçak et al., 1967); these observations having been confirmed using thin-section electron microscopy (Mendes Carneiro, 1975), which showed that paring also occurs in other regions of the homologous chromosomes. Electron microscope studies of the ultrastructure of chromatin fibers in Odontophrynus and ophidians have shown that the organization in these organisms is similar to that occurring in mammals in terms of the presence of nucleosomes, solenoids and higher levels of compaction (Beçak et al., 1977; Beçak and Fukuda, 1979). In 4n species, electron microscopy has also shown amplification of rDNA cistrons in pre-vitellogenic oocytes and, to a smaller degree, in pachytene spermatocytes of 4n species (Beçak et al., 1978; Beçak and Fukuda-Pizzocaro, 1980), while optical microscopy of 4n species has also shown AgAs banding in spermatocytes during the ‘bouquet’ phase (Beçak et al., 1978). The structure of chromosomes and the organization of DNA sequences have been discussed by Birnstein (1982) in a wide-ranging revision, which included anurans, urodela, reptiles, birds, and mammals. This study showed that there are great differences in the total quantity of DNA between Anura and Urodela, due to an increase in moderate repetitive DNA in the latter. Except for Xenopus mulleri, in anurans, unlike urodelans, satellite DNA was not found, and anurans possess a higher degree of methylation of genomic DNA and a larger number of palindromes. Comparison of the structure of amniotes and anamniotes suggests that the difficulty in obtaining G-banding in amphibians may be due to the higher degree of compaction of the chromatin fibers or to differences in the distribution of some DNA sequences. Birnstein (1982) suggested that fish and amphibians are phylogenetically closer than reptiles, birds and mammals, with reptiles being intermediate between the cold and warm-blooded groups. This was confirmed by Beçak et al. (1988), who used restriction-enzyme banding in Odontophrynus, and showed that the difficulty in obtaining G-bands is a result of the lower number of CG sequences (probably arranged in small agglomerations), or due to greater compaction of the chromatin, as had been proposed by various authors (Schmid, 1978; Birnstein, 1982; Bernardi et al., 1985; Schmid and Almeida, 1988). A closer phylogenetic relationship between reptiles and birds has been suggested by analytical centrifugation studies of DNA from the two groups, both of which present a CG-rich band of DNA, possibly located on microchromosomes (Comings et al., 1973). SILENCING OF REDUNDANT COPIES IN ODONTOPHRYNUS Research on tetraploid species has tried to clarify whether or not the greater genetic variability seen in these species has been generated by the greater gene complement, or due to alterations in gene regulation mechanisms, or both. It is known that in diploid and tetraploid O. americanus, total genome duplication causes no increase in total RNA content (Beçak and Goissis, 1971), even though tetraploids have double the number of 18S and 28S ribosomal genes, when compared to diploids (Schmidtke et al., 1976). The fact that ribosomal genes were not eliminated in 4n animals had been confirmed by the NOR studies of Ruiz et al. (1981). The quantitative synthesis of lactate dehydrogenase (LDH) and hemoglobin is reduced in tetraploid O. americanus, similar to what has been found in cryptic diploid species (Beçak and Pueyo, 1970). In an investigation of triploid and tetraploid Ceratophrydidae amphibians, Batistic et al. (1973) used autoradiography to demonstrate that although there were differences in satellite replication time between the parental chromosomes of triploid interspecific hybrids, asynchronous DNA replication does not occur in tetraploids. Although the gene activity of tetraploids is the same as in diploids, tetrasomic inheritance, as a result of genomic duplication, has led to greater genetic variability. This is supported by work on O. americanus, in which it has been shown that both for an albumin-type protein (Beçak et al., 1968b) and for tetrazolium oxidase (Schwantes et al., 1976, 1977) there are three phenotypes in diploids, while there are five in tetraploids. These data demonstrate that the sequences and types of heterozygotes occurring in tetraploid amphibians result in greater genetic polymorphism, and correlate with an increase in geographic distribution and greater variability in the ecological niches occupied by these animals (Beçak and Beçak, 1974a). It has also been found that diploid Hyla chrysoscelis and tetraploid Hyla versicolor have gene regulation similar to that described for diploid and tetraploid Odontophrynus, with the loss of non-essential DNA occurring in tetraploid Hyla (Bachmann and Bogart, 1975). Conjugated AgAs banding patterns and in situ rDNA hybridization data indicate a different behavior for one of the three nucleolar chromosomes of a triploid interspecific Odontophrynus hybrid (presumed to be between diploid O. cultripes and tetraploid O. americanus); the low level of transcription or silencing of the NOR region of this homologue being attributed to the smaller number of sequences or differences in the quantity of specific NOR-binding proteins in the triploid (Ruiz et al., 1980). Reduced genetic activity in the tetraploid could be at the transcription level and correlated to the higher level of methylation of the ribosomal genes, as suggested for the tetraploid O. americanus (Ruiz and Brison, 1989). These conclusions were supported by data on erythropoiesis and the transcription of DNA coding for hemoglobin in diploid and tetraploid O. americanus, which showed that tetraploid cells have only 30% more hemoglobin and 25-30% more ribosomes than diploid cells. Excess of Heinz bodies associated with RNA in RNP in tetraploid cells as compared to diploid cells may also reflect a system of regulation in which the excess mRNA transcribed in tetraploids is accumulated instead of being transcribed (Cianciarullo et al., 2000). The molecular organization of a-globlin genes in diploid and tetraploid O. americanus has shown that intron 2 is absent in both cases, indicating that these sequences may be pseudogenes related to retrotransposition (Acedo et al., 1997). Cloning and sequencing techniques for the study of ribosomal intergenic spacers (IGSs) have demonstrated a high degree of amplification of these regulatory sequences in O. americanus tetraploids, and it is also possible that transposon-like sequences have been inserted into these IGSs during evolution (Alvares et al., 1998). An epigenetic character found in Odontophrynus was the presence of amphiplasty. This was characterized by structural alterations in the genome involving differential condensation of the chromosomes or different expression of secondary restrictions. The fact that the two halves of the genome are morphologically dephased in different phases in the cell cycle would mean differences in the replication time of DNA, possibly related to methylation (Beçak and Beçak, 1998) (Figure 3). This cytological study found fragility in the secondary constriction of the fourth pair, paralleled by aneuploidies, in 2n and in 4n. The fragility of this locus was explained by an asynchrony in replication. The aneuploidies were attributed to a possible mutation involving the centromere, resulting in an epigenetic effect. It was also suggested that a mutation affected the dynamics of the centrosomes and of fibrillar fusion. Understanding this mutation is relevant, due to the involvement of centrosomes and of fibrillar fusion in neoplastic aneuploidies in humans (Borel et al., 2002).
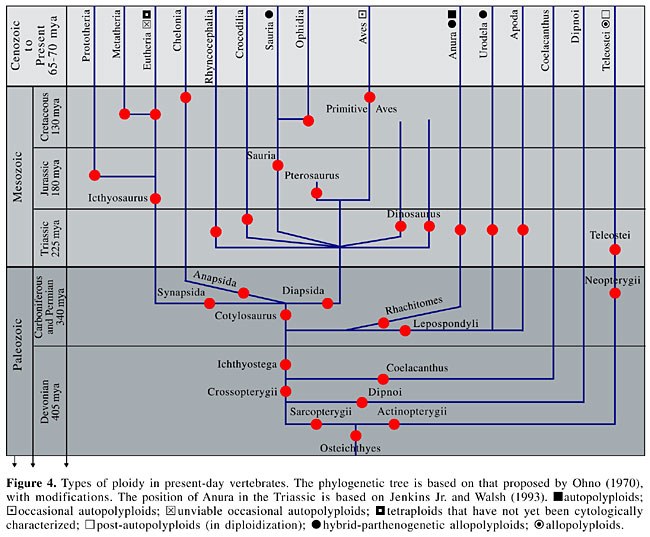
The importance of certain sequences of repetitive DNA for gene regulation processes in mammals, such as the inactivation of chromosome X (Lyon, 1998, 2000; Bailey at al., 2000) and imprinting mechanisms (Bird, 2002) has been emphasized. The compacting of heterochromatin, caused by highly repetitive sequences of DNA, is related to gene repression. The silencing of transcription would be through methylation, which in turn would be mediated by the acetylation of nucleosomal histones. In general, active loci are hyperacetylated and inactive loci are hypoacetylated (Strahl and Allis, 2000; Dillon and Festenstein, 2002; Hashimshony et al., 2003). Based on this line of thinking, certain alterations in methylation and in histone structure are transmitted through cell division, without changes in DNA sequences. These alterations that affect gene expression are known as “imprinting” (Park and Pfeifer, 2003). Alterations of this type are known from human embryonic development and they cause anomalies in parthenogenic and androgenetic embryos (Mutter et al., 1993). Evolution research has shown that the appearance and radiation of vertebrates were accompanied by the evolution of mechanisms of gene regulation through the repression of transcription. These events required an increase in methylation, accompanied by increases in methyltransferases (DNMT) and in the proteins that bind to CpG sites (Bestor, 1990; Bird, 1995). Consistent with this idea, the invertebrate Ciona intestinalis has only one gene for the protein MBD2/3, while the vertebrates have two genes, Mbd2 and Mbd3, probably originated through duplication (Hendrich and Tweedie, 2003). Loss of Dnmt1 causes alterations in gene expression, such as control of the cell cycle and the mobilization of retroelements (Jackson-Grusby et al., 2001). Abnormal hypermethylation can silence human tumor suppressor genes, which has lead to the idea that DNA methyltransferases have therapeutic properties (Baylin and Herman, 2000). As regards the role of histones in genetic regulation, there have been few studies on chromatin fiber ultrastructure and on the molecular organization of these proteins in amphibians and snakes (Birnstein, 1982). The chromatin fiber ultrastructure of these groups (Beçak et al., 1977; Beçak and Fukuda, 1979) is similar to that of mammals, in terms of nucleosomal organization and condensation into solenoids and other more condensed structures. It is known that in nucleosomal fiber, histone H1 binds to internucleosomal DNA, different from histones H2A, H2B, H3, and H4, which make up the central part of the nucleosome, which is wrapped with two 150-pb windings (Felsenfeld, 1992). It was also found that histone H1 has a regulatory role (Jost and Hofsteenge, 1992), preferentially binding to methylation sites containing CpG dinucleotides. In general, the regulatory proteins bind preferentially to the DNA palindromes (Mezquita et al., 1985). A comparative study of five species of vertebrates (trout, chickens, humans, rats, and mice) showed that the amino and carboxyl terminal regions of H1 have long palindromes formed by a succession of smaller ones. Long palindromes were not found in the other nucleosomal histones, or in the other regulatory proteins. Considerably variability was also found in the terminal regions of H1, and this variability was greater between closely related species, when compared to more distantly related species (Ohno and Beçak, 1993). The real role of H1 in gene expression has been debated (Hashimshony et al., 2003). In conclusion, the stability of the polyploid condition of Anura is a consequence of equilibrium of the processes that control chromosome dynamics during cellular division. The evolutionary flexibility is related to intra- and inter-populational chromosomal polymorphisms. The combination of gene dose (number of copies of a gene) and transcription control has epigenetic effects on genetic regulation. This conclusion is based on data on the activity of various isozymes, hemoglobin expression, silencing of ribosomal genes, nucleolar dominance in artificial hybrids, post-polyploidy amplification of repetitive sequences in the ribosomal spacer regions, the presence of transposons in these IGSs, and amphiplasty. EVOLUTION THROUGH POLYPLOIDY IN THE CHORDATA - HYPOTHESIS 2R The findings of autopolyploidy in anurans and post-polyploid fishes suggested the evolutive theory of polyploidy (Ohno, 1970, 1999). Based on this idea, the vertebrate genome evolved through two genomic duplications (2R), one in the transition of the invertebrates to the vertebrates, and the other in the divergence of the Osteichthyes (Figure 4). Consequently, the ancestors of the mammals, birds and reptiles, passed through tetraploidy at the fish or the amphibian phase. This hypothesis proposes that polyploidy furnished the raw material necessary for evolution, through the production of genetic redundancy. Posterior alterations of the copies would result in the acquisition of new functions, and in speciation. This hypothesis is supported by recent molecular findings, such as homeotic genes (Hox), in the field of developmental genetics. These genes, which promote the orientation and the differentiation of the anterior-posterior axis of the embryonal body, are only found in single doses in Amphioxus, while in the mammals there are four orthologs (Garcia-Fernandez and Holland, 1994). The variability of sequences between cDNA clones coding for SNAP-25, a synapse protein, is another demonstration of hypothesis 2R in fish (Larhammar and Risinger, 1994). Following these findings, various investigations have been made to find out when duplications occurred and to determine whether the increase in the genes is responsible for increased morphological complexity. The demonstration that the Hox clusters occur in the teleost Danio rerio, and in humans, revealed that these genes are anterior to the diversification of the Actinopterygii and Sarcopterygii (Postlethwait et al., 1998). The mapping of the teleost Fugu rubripes showed the occurrence of four Hox clusters, but with different characteristics. Three clusters are orthologous to those of the tetrapods, though genetic losses followed during evolution. The fourth cluster, HoxD, is not found in the tetrapods, and is apparently derived from an additional duplication (Aparicio et al., 1997). The “extra-” duplication was confirmed by the finding of seven Hox clusters in Danio rerio (Amores et al., 1998). The genomes of the mammals, and of the Coelacanth Latimeria menadoensis, which is derived from the genomic stock of the crossopterygians, show similarities in the Hox groupings; however, the Coelacanth has an additional Hox C1 gene, orthologous to Danio rerio, which was lost in the evolution of the mammals (Koh et al., 2003).
Comparative studies of the presence and expression of Hox genes have been made in the invertebrates. It was found that the branchiopod crustacean Artemia franciscana has at least four different Hox genes, which are equivalent to those of the insects (Averof and Akam, 1995). It was also found that certain genetic defects due to the Deformed gene, Dfd, of Drosophila melanogaster are remediated by the transfection of the human HOX 4.2 gene (McGinnis et al., 1990). Lynch and Conery (2000) proposed that the process of genomic duplication contributed more to the diversification of the species through the loss or silencing of extra-copies, than through the creation of new functions. This model is called speciation through duplication of the genome, and divergent resolution. The evolution of the duplicated genes can occur through three different routes: 1) a copy can be silenced by degenerative mutations (nonfunctionalization); 2) a copy can acquire a new function and be preserved through natural selection, while the other continues with the original function (“neo-functionalization”); 3) both copies could have mutations that reduce expression to that of the single-copy ancestor gene (“subfunctionalization”). Previously studies had been made of the electrophoretic patterns of an albumin type plasmatic protein, comparing the diploid anuran O. cultripes and the tetraploid O. americanus. While electrophoresis of the 2n, with two co-dominant alleles, showed three phenotypes in the population, following a (p + q)2 distribution, the 4n had five phenotypes, giving a (p + q)4 distribution (Beçak et al., 1968b; Beçak, 1969). Similar observations were found in the electrophoresis of the G6PD, 6-PGD and LDH enzyme patterns (Schwantes et al., 1969). The results demonstrated that the four genes of 4n were expressed in each individual, but quantitatively the final product was similar to that of the 2n. Two hypotheses were formulated (Beçak, 1969): 1) there was asynchrony in the genomes of the 4n, resulting in a situation in which one diploid lot was active at a time; 2) at the level of regulatory genes, there was a greater repression of the 4n genome, in comparison with the 2n genome that actually could be translated as a higher methylation level in the 4n as compared to the 2n. These two hypotheses could be correlated. These hypotheses approximate those of the mechanism proposed by Lynch and Conery (2000), called “subfunctionalization”. The suggestion that the tandem duplications also had an important role in the increase in anatomical complexity was reinforced by the comparative data of the Hox genes in Coelenterata and Bilateria (Rosa et al., 1999). An increase was found in the median Hox genes in the divergence of the Coelenterata and Bilateria, before the radiation of the latter group. Increases in the posterior Hox genes also would have occurred in the Enterocoela, including the vertebrates, after divergence from the other Bilateria. The theory of evolution through genomic duplication establishes that polyploidy ceases to occur from the reptiles on, due to the development of differentiated sexual chromosomes. The total duplication of the chromosomes would affect the equilibrium of the mechanism of sex determination (Figure 4). In the case of bisexual homeothermic animals, there are rare reports of autopolyploidy, as has been described in a 3n chicken (Ohno et al., 1963). Among the rodents, it was suggested that Mesocricetus auratus (2n = 44) is an allopolyploid species originated from the hybridization of Cricetus cricetus with Cricetus griseus (2n = 22). However, the DNA values (Moses and Yerganian, 1952) do not support this idea. The recent description of tetraploid rodents Tympanoctomys barrerae (Octodontidae, 4n = 102) is supported by DNA content data (Gallardo et al., 1999). Detailed cytological analyses remain to be made for a characterization of the type of ploidy. In humans, there are known cases of ploidies in fetuses, in inviable newborns, and in a few isolated cases of tetraploid or mosaic children, which have survived a few years (Edwards et al., 1967; Gardner, 1982). PANANIMALIA GENOME - CAMBRIAN Anaximander, a Greek philosopher (6th century B.C.) suggested that life began from a primordial mud, going through a sequence from inferior to superior life forms, giving rise to man from a type of fish. The notable paleontological discovery of the Cephalochordata Yunnanozoon lividum in Cambrian rocks in China (Chen et al, 1995) indicates that the Chordata emerged during the Cambrian explosion. This event apparently occurred during a short period of 10 million years. Equally interesting was the finding of the Agnatha Promissum pulchrum in the upper Ordovician in South Africa (Gabbot et al, 1995). According to Gould (1995), the Cambrian explosion indicates the abrupt formation of the Animal kingdom, in which all of the structural archetypes arose during the same period. The paleontological data, together with the molecular information of the Hox genes in animals that descended from the Cambrian, convinced Ohno (1996, 1999) that the second polyploidy coincided with the development of the Gnathostomata in the Ordovician period. The theory of the pananimalia genome proposed that various phyla of the animal kingdom emerged simultaneously during this short period of Cambrian explosion, without the individual genes presenting functional diversification. Otherwise, all of the animals of the various phyla would have the same genome, differing in the differential use of groups of individual genes (Ohno, 1996, 1997). A prototype would be Hallucigenia sparsa, probably having only one Hox gene, or a group of eight Hox genes, as in the arthropods. PHYLOGENY OF THE ANURANS BASED ON HYPOTHESIS 2R The phylogeny of the amphibia is not well resolved due to the rarity of fossils between the Paleozoic and the present. Based on the genetic theory of evolutive polyploidy, the origin of the modern amphibians (Anura, Urodela and Apoda) is polyphyletic (Ohno, 1970). The most primitive amphibian, Ichthyostega, of the extinct subclass Stegocephalia, would have appeared in the Devonian, through the evolution of Osteolepis fish (Crossopterygii, Rhipidistia) (Figure 4). In the following period, Carboniferous and Permian, these first amphibians, Ichthyostega, originated two phylogenetic lines, producing the Lepospondyli and the Rhachitomes. The former would have given origin to the modern Urodela and Gymnophiona, and the latter to the modern Anura. There would also have been branching in the Ichthyostega line, which gave rise to the first reptiles (Cotylosaurus). These reptiles would have given rise, through branching, to the Diapsida, Anapsida and Synapsida. The Diapsida branched out, giving rise to the dinosaurs, pterosaurs, other reptiles, birds, crocodiles, and Rhynchocephalia. The Anapsida apparently evolved to turtles, and the Synapsida became the Icthiosaurs and mammals. The molecular estimates, based on differences in gene sequences, were found to be coherent with calculations made based on fossil evidence. A difference between the two calculations was found in the diversification of certain orders of placental mammals, at the beginning of the Cretaceous, and before the extinction of the dinosaurs, in the Cretaceous-Jurassic period. A four times greater divergence in the molecular clock, than that previously established from fossil evidence, was also found for the rodents (Kumar and Hedges, 1998). Morescalchi, 1973, established a phylogenic model in Amphibia, comparing their data on DNA quantity and cytogenetic characters with those of various other authors. In Anura, they took into account the presence of microchromosomes in ancient families, such as the Ascaphidae and the Discoglossidae (Bogart, 1970), the polyploidy in the Leptodactylidae (Beçak et al., 1966, 1967; Bogart, 1967), centric fusions (Wickbom, 1945, 1949; Beçak, 1968; Rabello, 1970), the types of meiotic chiasmata in various families and the supernumerary chromosomes (Ullerich, 1967; Rabello, 1970). Single or multiple post-polyploidy translocations (Beçak and Beçak, 1974b, 1998) and translocations in diploids (Lourenço et al., 2000) were found after this analysis, as were differentiated sexual chromosomes (Schmid, 1980; Schempp and Schmid, 1981; Schmid et al., 1983). The model of Morescalchi indicates that in Anura the oldest families, Ascaphidae and Discoglossidae, evolved to Pipidae, Rhinophrynidae, Pelobatidae, and Leptodactylidae, in the upper Jurassic. Hylidae, Ranidae and Bufonidae originated from the Leptodactylidae. Fossils of the Microhylidae have only been found in the Cenozoic (Miocene); it is not known if these anurans arose recently, or if they are more primitive. We conclude that the genomic duplication in tetraploid and octoploid anurans in the Leptodactylidae, Hylidae and other families, indicates that this mechanism can occur as an independent event, and more than once in the same evolutionary lineage. ACKNOWLEDGMENTS We thank Helir Serralvo for help with editing and with photomicroscopy. REFERENCES Acedo, M.D.P., Paranhos-Baccalà, G., Denoya, C.D. and Ruiz, I.R.G. (1997). Molecular cloning of exons II and III of the a-globin major gene from Odontophrynus americanus 2n and 4n (Amphibia, Anura). Braz. J. Genet. 20: 613-617. Adams, K.L., Cronn, R., Percifield, R. and Wendel, J.F. (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100: 4649-4654. Almeida, T.M.B., Ruiz, I.R.G. and Beçak, W. (1986). Ribosomal gene activity detected by silver staining in two diploid populations of Odontophrynus americanus (Amphibia, Anura) from Southern Brazil. Rev. Bras. Genet. IX: 433-437. Alvares, L.E., Brison, O. and Ruiz, I.R.G. (1998). Identification of enhancer-like elements in the ribosomal intergenic spacer of Odontophrynus americanus 2n and 4n (Amphibia, Anura). Genetica 104: 41-44. Amores, A., Force A., Yan, Y.-L., Joly, L., Amemiya, C., Fritz, A., Ho, R.K., Langeland, J., Prince, V., Wang, Y.-L., Westerfield, M., Ekker, M. and Postlethwait, J.H. (1998). Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711-1714. Aparicio, S., Hawker, K., Cottage, A., Mikawa, Y., Zuo, L., Venkatesh, B., Chen, E., Krumlauf, R. and Brenner, S. (1997). Organization of the Fugu rubripes Hox clusters: evidence for continuing evolution of vertebrate Hox complexes. Nat. Genet. 16: 79-83. Atkin, N.B. and Ohno, S. (1967). DNA values of four primitive chordates. Chromosoma 23: 10-13. Averof, M. and Akam, M. (1995). Hox genes and the diversification of insect and crustacean body plans. Nature 376: 420-423. Bachmann, K. and Bogart, J.P. (1975). Comparative cytochemical measurements in the diploid-tetraploid species pair of hylid frogs Hyla chrysoscelis and H. versicolor. Cytogenet. Cell Genet. 15: 186-194. Bailey, J.A., Carrel, L., Chakravarti, A. and Eichler, E.E. (2000). Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc. Natl. Acad. Sci. USA 97: 6634-6639. Batistic, R.F., Beçak, W. and Beçak, M.L. (1973). DNA autoradiographic patterns in diploid, triploid and tetraploid amphibians (Ceratophrydidae). Cytologia 36: 687-697. Batistic, R.F., Soma, M., Beçak, M.L. and Beçak, W. (1975). Further studies on polyploid amphibians - A diploid population of Phillomedusa burmeisteri. J. Hered. 66: 160-162. Baylin, S.B. and Herman, J.G. (2000). DNA hypermethylation in tumorigenesis. Trends Genet. 16: 168-173. Beçak, M.L. (1968). Chromosomal analysis of eighteen species of Anura. Caryologia 21: 191-208. Beçak, M.L. and Beçak, W. (1970). Further studies on polyploid amphibians (Ceratophrydidae). III. Meiotic aspects of the interspecific triploid hybrid: Odontophrynus cultripes (2n = 22) x O. americanus (4n = 44). Chromosoma 31: 377-385. Beçak, M.L. and Beçak, W. (1974a). Studies on polyploid amphibians. Karyotype evolution and phylogeny of the genus Odontophrynus. J. Herpetol. 8: 337-341. Beçak, M.L. and Beçak, W. (1974b). Diploidization in Eleutherodactylus (Leptodactylidae-Amphibia). Experientia 30: 624-625. Beçak, M.L. and Beçak, W. (1998). Evolution by polyploidy in Amphibia: new insights. Cytogenet. Cell Genet. 80: 28-33. Beçak, M.L. and Fukuda, K. (1979). Arrangement of nucleosomes in condensed chromatin fibres. Experientia 35: 24-26. Beçak, M.L. and Fukuda-Pizzocaro, K. (1980). Chromatin circles in amphibian previtelogenic oocytes. Experientia 36: 164-166. Beçak, M.L., Beçak, W. and Rabello, M.N. (1966). Cytological evidence of constant tetraploidy in the bisexual South American frog Odontophrynus americanus. Chromosoma 19: 188-193. Beçak, M.L., Beçak, W. and Rabello, M.N. (1967). Further studies on polyploid amphibians (Ceratophrydidae). I. Mitotic and meiotic aspects. Chromosoma 22: 192-201. Beçak, M.L., Denaro, L. and Beçak, W. (1970a). Polyploidy and mechanisms of karyotypic diversification in Amphibia. Cytogenetics 9: 225-238. Beçak, M.L., Beçak, W. and Vizotto, L.D. (1970b). A diploid population of the polyploid amphibian Odontophrynus americanus and an artificial intraspecific triploid hybrid. Experientia 26: 545-546. Beçak, M.L., Fukuda, K. and Mendes Carneiro, S. (1977). Chromatin ultrastructure of lower vertebrates. Experientia 33: 1314-1316. Beçak, M.L., Mendes Carneiro, S. and Fukuda, K. (1978). Circles in spermatocyte chromatin loops. Electron microscopy and AgAs-NORs studies. Experientia 34: 171-172. Beçak, M.L., Stocco dos Santos, R.C., Soares-Scott, M.D., Batistic, R.F. and Costa, H. (1988). Chromosome structure in man and Amphibia-Anura, restriction enzymes. Rev. Bras. Genet. 11: 939-948. Beçak, W. (1969). Genic action and polymorphism in polyploid species of amphibians. Genetics 61 (Suppl): 183-190. Beçak, W. and Goissis, G. (1971). DNA and RNA content in diploid and tetraploid amphibians. Experientia 27: 345-346. Beçak, W. and Pueyo, M.T. (1970). Gene regulation in the polyploid amphibian Odontophrynus americanus. Exp. Cell Res. 63: 448-451. Beçak, W., Beçak, M.L., Lavalle, D. and Schreiber, G. (1967). Further studies on polyploid amphibians (Ceratophrydidae) II. Content and nuclear volume. Chromosoma 23: 14-23. Beçak, W., Beçak, M.L. and Langlada, F.G. (1968a). Artificial triploid hybrids by interspecific mating of Odontophrynus (Amphibia-Anura). Experientia 24: 1162-1163. Beçak, W., Schwantes, A.R. and Schwantes, M.L. (1968b). Polymorphism of albumin-like proteins in the South American tetraploid frog Odontophrynus americanus (Salientia: Ceratophrydidae). J. Exp. Zool. 168: 473-476. Beçak, W., Beçak, M.L., Schreiber, G., Lavalle, D. and Amorim, F.O. (1970). Interspecific variability of DNA content in Amphibia. Experientia 22: 204-206. Bernardi, G., Olofsson, B., Filipski, J., Zerial, M., Salinas, J., Cuny, G., Meunier-Rotival, M. and Rodier, F. (1985). The mosaic genome of warm-blooded vertebrates. Science 228: 953-958. Bestor, T.H. (1990). DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Philos. Trans. R. Soc. Lond.B. Biol. Sci. 326: 179-187. Bird, A.P. (1995). Gene number, noise reduction and biological complexity. Trends Genet. 11: 94-100. Bird, A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16: 6-21. Birnstein, V.J. (1982). Structural characteristics of genome organization in Amphibia: Differential staining of chromosomes and DNA structure. J. Mol. Evol. 18: 73-91. Bogart, J.P. (1967). Chromosomes of the South American amphibian family Ceratophridae with a reconsideration of the taxonomic status of Odontophrynus americanus. Can. J. Genet. Cytol. 9: 531-542. Bogart, J.P. (1970). Systematic problems in the amphibian family Leptodactylidae (Anura) as indicated by karyotypic analysis. Cytogenetics 9: 369-383. Bogart, J.P. and Wasserman, A.O. (1972). Diploid-polyploid cryptic species pairs: a possible clue to evolution by polyploidization in anuran amphibians. Cytogenetics 11: 7-24. Borel, F., Lohez, O.D., Lacroix, F.B. and Margolis, R.L. (2002). Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc. Natl. Acad. Sci. USA 99: 9818-9824. Chen, J.-Y., Dzik, J., Edgecombe, G.-D., Ramsköld, L. and Zhou, G.-Q. (1995). A possible Early Cambrian chordate. Nature 377: 720-722. Cianciarullo, A.M., Naoum, P.C., Bertho, A.L., Kobashi, L.S., Beçak, W. and Soares, M.J. (2000). Aspects of gene regulation in the diploid and tetraploid Odontophrynus americanus (Amphibia, Anura, Leptodactilydae). Gen. Mol. Biol. 23: 357-364. Comings, D.E., Avelino, E. and Beçak, W. (1973). Heavy shoulder DNA in snakes. Cytogenet. Cell Genet. 12: 2-7. Cortadas, J. and Ruiz, I.R.G. (1988). The organization of ribosomal genes in diploid and tetraploid species of the genus Odontophrynus (Amphibia, Anura). Chromosoma 96: 437-442. Dillon, N. and Festenstein, R. (2002). Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 18: 252-258. Edwards, J.H., Yuncken, C., Rushton, D.I., Richards, S. and Mittwoch, U. (1967). Three cases of triploidy in man. Cytogenetics 6: 81-104. Felsenfeld, G. (1992). Chromatin as an essential part of the transcriptional mechanism. Nature 355: 219-224. Gabbot, S.E., Aldridge, R.J. and Theron, J.N. (1995). A giant conodont with preserved muscle tissue from the upper Ordovician of South Africa. Nature 374: 800-803. Gallardo, M.H., Bickham, J.W., Honeycutt, R.L., Ojeda, R.A. and Köhler, N. (1999). Discovery of tetraploidy in a mammal. Nature 401: 341. Garcia-Fernandez, J. and Holland, P.W. (1994). Archetypal organization of the amphioxus Hox genes cluster. Nature 370: 563-566. Gardner, L.I. (1982). The lessons of polyploid. Relation to congenital asymmetry and the Russell-Silver syndrome. Am. J. Dis. Child. 36: 292-293. Gould, S.J. (1995). Of it, not above it. Nature 377: 681-682. Hashimshony, T., Zhang, J., Keshet, I., Bustin, M. and Cedar, H. (2003). The role of DNA methylation in setting up chromatin structure during development. Nat. Genet. 34: 187-192. Hendrich, B. and Tweedie, S. (2003). The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 19: 269-277. Jackson-Grusby, L., Beard, C., Possemato, R., Tudor, M., Fambrough, D., Csankovszki, G., Dausman, J., Lee, P., Wilson, C., Lander, E. and Jaenisch, R. (2001). Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27: 31-39. Jenkins Jr., F.A. and Walsh, D.M. (1993). An Early Jurassic caecilian with limbs. Nature 365: 246-250. Jost, J.P. and Hofsteenge, J. (1992). The repressor MDBP-2 is a member of the histone H1 family that binds preferentially in vitro and in vivo to methylated nonspecific DNA sequences. Proc. Natl. Acad. Sci. USA 89: 9499-9503. Kawamura, T. (1984). Polyploidy in amphibians. Zool. Sci. 1: 1-5. Koh, E.G.L., Lam, K., Christoffels, A., Erdmann, M.V., Brenner, S. and Venkatesh, B. (2003). Hox gene clusters in the Indonesian Coelacanth, Latimeria menadoensis. Proc. Natl. Acad. Sci. USA 100: 1084-1088. Kumar, S. and Hedges, S.B. (1998). A molecular time scale for vertebrate evolution. Nature 392: 917-919. Larhammar, D. and Risinger, C. (1994). Why so few pseudogenes in the tetraploid species? Trends Genet. 10: 418-419. Lourenço, L.B., Recco-Pimentel, S.M. and Cardoso, A.J. (2000). A second case of multivalent meiotic configurations in diploid species of Anura. Genet. Mol. Biol. 23: 131-133. Lynch, M. and Conery, J.S. (2000). The evolution fate and consequences of duplicate gene. Science 290: 1151-1155. Lyon, M.F. (1998). X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell Genet. 80: 133-137. Lyon, M.F. (2000). LINE-1 elements and X chromosome inactivation: A function for “junk” DNA? Proc. Natl. Acad. Sci. USA 97: 6248-6249. Martin, A.P. (1999). Increasing genomic complexity by gene duplication and the origin of vertebrates. Am. Nat. 154: 111-128. Mendes-Carneiro, S. (1975). Observações sobre a ultra-estrutura das células germinativas masculinas da espécie diplo-tetraplóide de Odontophrynus americanus (Amphibia-Anura). Mem. Inst. Butantan 39: 135-148. McGinnis, N., Kuziora M.A. and McGinnis, W. (1990). Human Hox-4.2 and Drosophila Deformed encode similar regulatory specificities in Drosophila embryos and larvae. Cell 63: 969-976. Mezquita, J., Connor, W., Einkfein, R.J. and Dixon, G.H. (1985). An H-1 histone gene from rainbow-trout (Salmo gairdnerii). J. Mol. Evol. 21: 209-219. Moore, G. (2002). Meiosis in allopolyploids - the importance of “Tefloon” chromosomes. Trends Genet. 18: 456-463. Morescalchi, A. (1973). Amphibia. In: Cytotaxonomy and Vertebrate Evolution (Chiarelli, A.B. and Capanna, E., eds.). Academic Press, London, New York, pp. 223-348. Moses, M.J. and Yerganian, G. (1952). Desoxypentose nucleic acid (DNA) content and cytotaxonomy of several Cricetinae (hamster). Genetics 37: 607-608. Muller, H.J. (1925). Why polyploidy is rare in animals than in plants. Ann. Nat. 59: 346-353. Mutter, G.L., Stewart, C.L., Chaponot, M.L. and Pomponio, R.J. (1993). Oppositely imprinted genes H19 and insulin-like growth factor 2 are coexpressed in human androgenetic trophoblast. Am. J. Hum. Genet. 53: 1096-1102. Naruya, S. (2002). Evolutionary genomics: molecular evolution at the genomic scale. Trends Genet. 18: 239-240. Ohno, S. (1970). Evolution by Gene Duplication. Spring-Verlag, Berlin, Heidelberg, New York. Ohno, S. (1996). The notion of the Cambrian pananimalia genome. Proc. Natl. Acad. Sci. USA 93: 8475-8478. Ohno, S. (1997). The reason for as well as the consequence of the Cambrian explosion in animal evolution. J. Mol. Evol. 44 (Suppl): S23-S27. Ohno, S. (1999). Gene duplication and the uniqueness of vertebrate genome circa 1970-1999. Sem. Cell Dev. Biol. 10: 517-522. Ohno, S. and Atkin, N.B. (1966). Comparative DNA values and chromosome complements of eight species of fishes. Chromosoma 18: 455-466. Ohno, S. and Beçak, M.L. (1993). Can a protein influence the fate of its own coding sequence? The amino- and carboxyl-terminal regions of H1 histone. Proc. Natl. Acad. Sci. USA 90: 7341-7345. Ohno, S., Kittrell, W.A., Christian, L.C., Stenius, C. and Witt, G.A. (1963). An adult triploid chicken (Gallus domesticus) with a left ovotestis. Cytogenetics 2: 42-49. Ohno, S., Wolf, U. and Atkin, N.B. (1968). Evolution from fish to mammals by gene duplication. Hereditas 59: 169-187. Osborn, T.C., Pires, J.C., Birchler, J.A., Auger, D.L., Chen, Z.J., Lee, H.S., Comai, L., Madlung, A., Doerge, R.W., Colot, V. and Martienssen, R.A. (2003). Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19: 141-147. Park, K.-Y. and Pfeifer, K. (2003). Epigenetic interplay. Nat. Genet. 34: 126-128. Postlethwait, J.H., Yan, Y.L., Gates, M.A., Horne, S., Amores, A., Brownlie, A., Donovan, A., Egan, E.S., Force, A., Gong, Z., Goutel, C., Fritz, A., Kelsh, R., Knapik, E., Liao, E., Paw, B., Ransom, D., Singer, A., Thomson, M., Abduljabbar, T.S., Yelick, P., Beier, D., Joly, J.S., Larhammar, D., Rosa, F., Westerfield, M., Zon, L.I., Johnson, S.L., Talbot, W.S. and Ekker, M. (1998). Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 18: 345-349. Rabello, M.N. (1970). Chromosomal studies in Brazilian anurans. Caryologia 23: 45-59. Rosa, R. de, Grenier, J.K., Andreeva, T., Cook, C.E., Adoutte, A., Akam, M., Carroll, S.B. and Balavoine, G. (1999). Hox genes in brachiopods and priapulids and protostome evolution. Nature 339: 772-776. Ruiz, I.R.G. and Beçak, W. (1976). Further studies on polyploid amphibians V. C-banding in diploid and tetraploid species of Odontophrynus. Chromosoma 54: 69-74. Ruiz, I.R.G. and Brison, O. (1989). Methylation of ribosomal cistrons in diploid and tetraploid Odontophrynus americanus (Amphibia, Anura). Chromosoma 98: 86-92. Ruiz, I.R.G., Bonaldo, M.F. and Beçak, W. (1980). In situ localization of ribosomal genes in a natural triploid of Odontophrynus. J. Hered. 71: 55-57. Ruiz, I.R.G., Soma, M. and Beçak, W. (1981). Nucleolar organizer regions and constitutive heterochromatin in polyploid species of the genus Odontophrynus (Amphibia, Anura). Cytogenet. Cell Genet. 29: 84-98. Saez, F.A. and Brum-Zorilla, N. (1966). Karyotype variation in some species of the genus Odontophrynus (Amphibia, Anura). Caryologia 19: 55-63. Schempp, W. and Schmid, M. (1981). Chromosome banding in Amphibia. VI. BrdU-replication patterns in anura and demonstration of XX/XY sex chromosomes in Rana esculenta. Chromosoma 83: 697-710. Schmid, M. (1978). Chromosome banding in Amphibia I. Constitutive heterochromatin and nucleolar regions in Bufo adn Hyla. Chromosoma 66: 361-388. Schmid, M. (1980). Chromosome banding in Amphibia. V. Highly differentiated ZZ/ZW sex chromosomes and exceptional genomes size in Pyxicephalus adspersus (Anura, Ranidae). Chromosoma 80: 69-96. Schmid, M. and Almeida, C.G. (1988). Chromosome banding in Amphibia XII. Restriction endonuclease banding. Chromosoma 96: 283-290. Schmid, M., Haaf, T., Geile, B. and Sims, S. (1983). Unusual heteromorphic sex chromosomes in a marsupial frog. Experientia 39: 1153-1155. Schmid, M., Haaf, T. and Schempp, W. (1985). Chromosome banding in Amphibia IX. The polyploid karyotypes of Odontophrynus americanus and Ceratophrys ornata (Anura, Leptodactylidae). Chromosoma 91: 172-184. Schmidtke, J., Beçak, W. and Engel, W. (1976). The reduction of genic activity in the tetraploid Odontophrynus americanus is not due to loss of ribosomal DNA. Experientia 32: 27-28. Schwantes, A.R., Schwantes, M.L.B. and Beçak, W. (1969). Electrophoretic patterns of G-6-PD, 6-PGD and LDH in polyploid amphibians (Ceratophrydidae). Rev. Bras. Pesqui. Med. Biol. 2: 41-44. Schwantes, M.L.B., Schwantes, A.R. and Beçak, W. (1976). Estudo comparativo de dez enzimas num sistema diploide do gênero Odontophrynus americanus (Ceratophrynidae-Anura). Cienc. Cult. 28 (Suppl): 280-281. Schwantes, M.L.B., Schwantes, A.R. and Beçak, W. (1977). Electrophoretic studies on polyploid amphibians. I. 6-phosphogluconatedehydrogenase (6-PGD). Comp. Biochem. Physiol. 56B: 393-396. Simmen, N.W., Leitgeb, S., Clarck, V.H., Jones, S.J.M. and Bird, A. (1998). Gene number in an invertebrate chordata, Ciona intestinalis. Proc. Natl. Acad. Sci. USA 95: 4437-4460. Soares-Scott, M.D., Trajtengertz, I., Soma, M. and Beçak, M.L. (1988). C and AgAs bands of the octaploid untanha frog Ceratophrys dorsata (C. aurita) (8n = 104, Amphibia, Anura). Rev. Bras. Genet. 11: 625-631. Spring, J. (1997).Vertebrate evolution by interspecific hybridisation - are we polyploid? FEBS Lett. 400: 2-8. Strahl, B. and Allis, C.D. (2000). The language of covalent histone modifications. Nature 403: 41-45. Ullerich, F. (1967). Weitere untersuchungen über chromosomen verhältnisse und DNS-gehalt bei Anuran (Amphibia). Chromosoma 21: 345-368. Wickbom, T. (1945). Cytological studies on Dipnoi, Urodela, Anura and Emys. Hereditas 31: 241-346. Wickbom, T. (1949). Further cytological studies on Anura and Urodela. Hereditas 35: 33-48. Wolf, V., Ritter, H., Atkin, N.B. and Ohno, S. (1969). Polyploidization in the fish family Cyprinidae, order Cypriniformes. I. DNA-content and chromosome sets in various species of Cyprinidae. Humangenetik 7: 240-244. |
|