ABSTRACT. Like all nitrosamines, N-nitrosodiethylamine (NDEA) requires metabolic activation in order to exert its carcinogenic effects. This activation involves cytochrome P450s (CYP), which generates unstable metabolites that react with the DNA of cells in the immediate vicinity of metabolite formation. Although NDEA is carcinogenic, it has been considered a weak mutagen in classic genotoxicity assays. We used optimized Salmonella/mammalian microsome genotoxicity assays to assess the mutagenicity and toxicity of low concentrations of NDEA. Using a fixed concentration of NDEA (36.5 mg/ml), we varied the length of preincubation in the presence of different concentrations of an S9 metabolic activation mixture. Salmonella typhimurium strains TA97 and TA102 were resistant to NDEA-induced mutagenesis, even after a preincubation of up to 120 min and the use of different concentrations of the S9 mix. Strain TA98 was susceptible to mutagenesis by NDEA in the absence of the S9 mix and after preincubation with NDEA for 90 min. When bacteria of this strain were preincubated with NDEA for 60 min, mutagenesis was detected at an S9 mix concentration >9.55 mg/ml. NDEA also induced mutagenesis in strain TA100 after preincubation for 90 or 120 min, and this effect was dependent on the S9 concentration. E. coli strain BH990 also showed a concentration-dependent response, with only 60% of the cells surviving after a 120-min preincubation with NDEA in the presence of 19.1 mg S9 mix/ml. Key words: Metabolic activation, N-nitrosodiethylamine, Preincubation, Survival curve INTRODUCTION Nitrosamines are precarcinogens capable of inducing tumors in different animal species and are suspected of being involved in some human tumors (Lijinsky, 1992). N-Nitrosodiethylamine (NDEA), which is present in the environment (Tricker and Preussmann, 1991) and in tobacco smoke, and is also synthesized endogenously (Sander, 1967), induces tumors in all species tested so far (Bogovski and Bogovski, 1981). Like all nitrosamines, NDEA requires metabolic activation in order to exert its carcinogenic effects (Bartsch and Montesano, 1984). This activation involves cytochrome P450s (CYP) which generate unstable metabolites with a very short half-life that react with the DNA of cells close to the sites of metabolite formation (Berger et al., 1987). The main premutagenic bases generated from NDEA are O6-ethylguanine and O4-ethylthymine (Pegg and Byers, 1992). These premutagenic bases are repaired by alkylguanine transferase, which removes the alkyl group from the modified base in a suicide fashion (Nakatsuru et al., 1993). The administration of NDEA to rats results in lipid peroxidation and enhanced chemiluminescence in liver preneoplastic nodules, indicating the formation of activated oxygen species (Pegg, 1983; Nakae et al., 1997). NDEA also produces 8-hydroxyguanine (8-OHG) (Ampy and Williams, 1986), an indicator of oxidative damage to DNA and the most abundant of more than 20 types of modifications produced under conditions of oxidative stress. This premutagenic DNA damage results in specific types of mutations and is likely to be involved in carcinogenesis. Although NDEA is carcinogenic, it is still considered to be a weak mutagen in classic genotoxicity assays (McCann et al., 1975; Maron and Ames, 1983; Zeiger et al., 1987). The reasons for this are not entirely understood, but probably reflect non-optimal experimental conditions, such as the absence or presence of certain enzymes that may interfere with the activation and/or repair of DNA lesions generated by NDEA. In the present study, we used optimized Salmonella/mammalian microsome genotoxicity assays to examine the responses to low concentrations of NDEA. MATERIAL AND METHODS Chemicals NDEA and the positive controls were obtained from Sigma Co. (St. Louis, MO, USA) and were dissolved in 0.9% NaCl (w/v) prior to use. Bacterial strains The Salmonella typhimurium strains TA98, TA97, TA100, and TA102 described by Maron and Ames (1983) were provided by Dr. B.N. Ames (University of California, Berkeley, CA, USA). The Escherichia coli strains CC104, BH990, BH980, and BH540 were provided by Dr. S. Boiteux (Department de Radiobiologie et Radiopathologie, Center National de la Recherche Scientifique, Fontenay-aux-Rose, France). The genetic characteristics of the strains used are listed in Table 1.
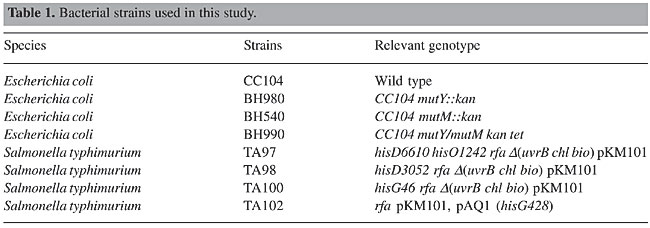
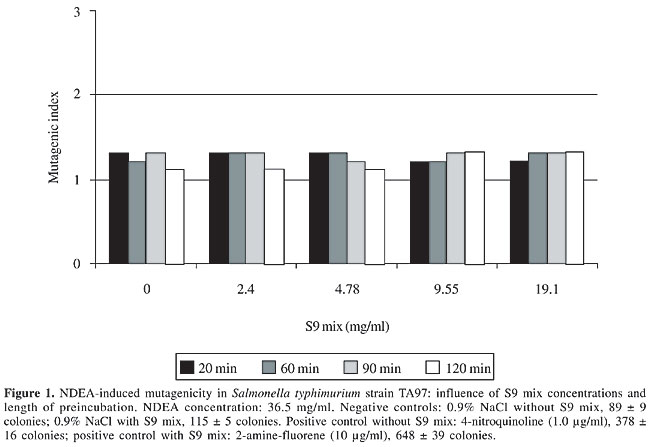
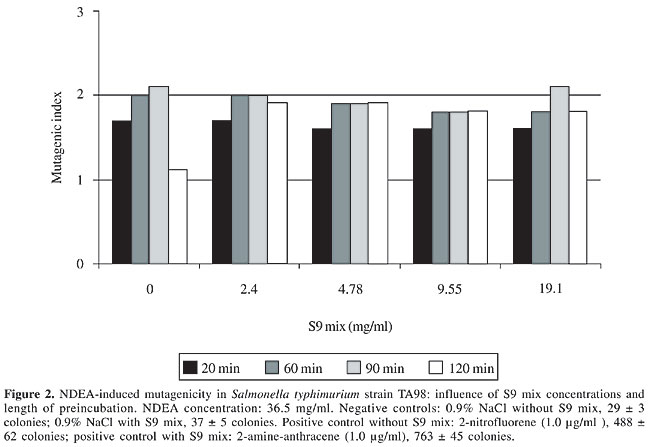
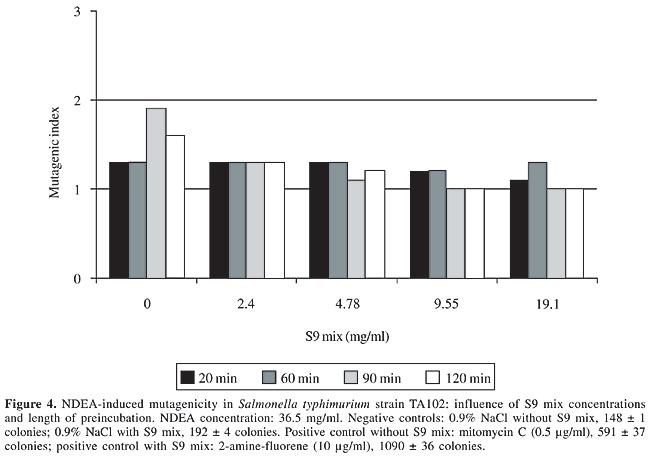
S9 fraction The S9 fraction prepared from the liver of Sprague-Dawley rats pretreated with a polychlorinated biphenyl mixture (Aroclor 1254) was purchased from Molecular Toxicology Inc. (MoltoxTM,USA). The S9 metabolic activation mixture (S9 mix) was prepared according to Maron and Ames (1983) and was used in Salmonella/microsome tests and survival assays as described elsewhere (da Costa Lopes et al., 2000), with modifications. Salmonella/mammalian microsome assay Mutagenicity was assessed using the procedure described by Maron and Ames (1983) with S. typhimurium strains TA98, TA97, TA100, and TA102 preincubated with 36.5 µg NDEA/ml, in the absence and presence of S9 mix. The assay mixture consisted of 100 µl NDEA solution, 500 µl S9 mix (2.4, 4.78, 9.55, and 19.1 mg total protein/ml) or the same volume of 0.2 M sodium phosphate buffer, pH 7.4, in experiments without S9 mix, and 100 µl bacterial suspension (2 x 109 cells/ml). The mixture was preincubated at 37°C with shaking. After 20, 60, 90 and 120 min, 2 ml top agar (0.6% agar, 0.6% NaCl w/v, 50 µM L-histidine, 50 µM biotin, pH 7.4, 45°C) was added to the test tube and the final mixture was poured onto a Petri dish with minimal agar (1.5% agar, Vogel-Bonner E medium, containing 2% glucose). This final mixture was incubated at 37°C for 72 h, and the His+ revertant colonies were counted. The results were expressed as a mutagenic index (MI = number of His+ induced in the sample/number of spontaneous His+ in the negative control). Positive controls for the assays in the absence or presence of S9 mix were done as described elsewhere (Aiub et al., 2003). NDEA was considered to be mutagenic when: a) the number of revertant colonies in the assay was at least twice the number of spontaneous revertants (MI ³2), b) analysis of variance (ANOVA) revealed a significant response (P £ 0.05), and c) a reproducible concentration-response curve (P £ 0.01) was obtained. This evaluation was based on previously established criteria (Vargas et al., 1993, 1995). Survival experiments To determine the cytotoxicity, cultures of each bacterial strain were preincubated in a final volume of 700 µl (100 µl 2 x 109 cells/ml, 100 µl NDEA (36.5 mg/ml) or 0.9% NaCl, and 500 µl of each concentration of S9 mix or 0.2 M sodium phosphate buffer, pH 7.4). After 60, 90 and 120 min, the cells were washed and diluted in 0.9% NaCl to obtain a suspension containing 2 x 103 cells/ml. A suitable aliquot (100 µl) of this suspension was plated on nutrient agar (0.8% bacto nutrient broth (Difco), 0.5% NaCl and 1.5% agar), after which the plates were incubated at 37°C for 24 h and the colonies then counted. Each experiment was done in triplicate and was repeated at least twice, with the standard deviation among replicates not exceeding 15%. Analysis of variance (ANOVA) was used for statistical comparisons, with a value of P £ 0.05 indicating a significant response (Vargas et al., 1993, 1995). RESULTS AND DISCUSSION In the present study, we examined the mutagenicity and genotoxicity of NDEA under conditions different from those normally used in these assays. This approach was necessary since NDEA is a complete carcinogen (Pegg, 1983), but has not consistently produced positive mutagenic responses in the usual Ames assay done under conventional conditions (McCann et al., 1975; Maron and Ames, 1983). The standard protocol in the Ames assay stipulates a preincubation of 20 min. This preincubation period is very important since it allows NDEA to be efficiently activated and ensures repair of the lesion produced in bacterial DNA since the bacterial mitotic cycle lasts around 30 min. Alternatively, some of the metabolites generated from the metabolic activation of NDEA may induce DNA damage that is not repaired, thereby resulting in cytotoxicity and subsequent cell death. To determine the best conditions for mutagenesis, we used several preincubation periods (20 to 120 min) and different concentrations of the S9 mix (2.4 to 19.1 mg/ml) while maintaining a low NDEA concentration (36.5 mg/ml). Figures 1 to 4 show the variation in mutagenesis for each S. typhimurium strain under these conditions. Strain TA97 was resistant to NDEA-induced mutagenesis, regardless of the concentration of S9 mix used or the length of preincubation (up to 120 min) (Figure 1). Strain TA98 was susceptible to NDEA-induced mutagenesis without the addition of S9 mix or when preincubated with NDEA for 90 min (Figure 2). When this strain was preincubated with NDEA for 60 min, mutagenesis was detected at S9 mix concentrations >9.55 mg/ml. NDEA caused concentration-dependent mutagenensis in strain TA100 after preincubation for 90 or 120 min (Figure 3), although only with the former preincubation were there positive results (the cytotoxicity caused by the 120-min preincubation gave a false-positive result). NDEA did not cause mutagenesis in strain TA102, even at an S9 mix concentration of 19.1 mg/ml or after a 120-min preincubation (Figure 4). Thus, NDEA was apparently unable to cause frameshift mutagenicity or A-T base pair reversions. Overall, these results suggested that the principal effects of the metabolic activation of NDEA involved G-C base pair reversions.
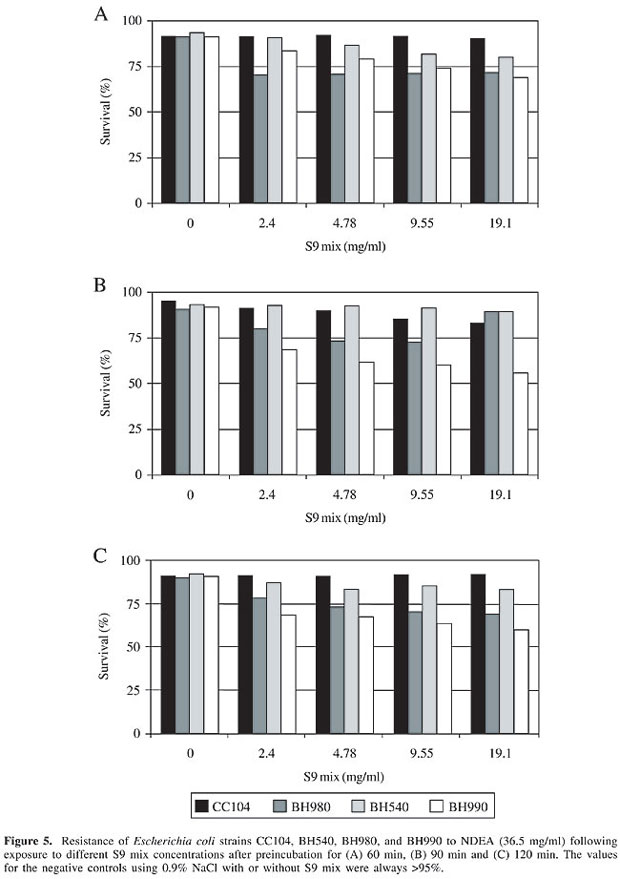
Cytotoxicity was detected in the E. coli strain BH990 (mutM/mutY, a double mutant) at all of the S9 mix concentrations tested after a 90-min preincubation. Cytotoxicity was also detected in BH980 (mutY) at an S9 concentration of 19.1 mg/ml after 60- and 90-min preincubations (Figure 5). These findings suggested that the cytotoxicity produced by the activation of NDEA resulted from an action on sugar residues and bases, and produced lesions that could be repaired by the base excision repair pathway. MutM and MutY DNA glycosylases cooperate in this repair system to eliminate either 8-OH or the misincorporated adenine opposite 8-OH, respectively (Tchou and Grollman, 1993).
We are currently developing new bacterial strains capable of expressing cytochrome P450s, such as CYP2E1 (Yoo et al., 1990) and CYP2A3 (Ribeiro Pinto et al., 2001), that can activate NDEA within these organisms and increase the chances that the electrophile metabolites generated from NDEA activation will react with DNA. The results of the present study indicate that the best strains for these transformations are TA98 and TA100. ACKNOWLEDGMENTS Research supported by FAPERJ, CNPq, SR2-UERJ, and Programa Prociência. REFERENCES Aiub, C.A.F., Ribeiro Pinto, L.F. and Felzenszwalb, I. (2003). N-Nitrosodiethylamine mutagenicity at low concentrations. Toxicol. Lett. 145: 36-45. Ampy, F.R. and Williams, A.O. (1986). Dimethylnitrosamine metabolism: I. In vitro activation of dimethylnitrosamine to mutagenic substance(s) by hepatic and renal tissues from three inbred strains of mice. Life Sci. 39: 923-930. Bartsch, H. and Montesano, R. (1984). Relevance of nitrosamines to human cancer. Carcinogenesis 5: 1381-1393. Berger, M.R., Schmähl, D. and Zerban, H. (1987). Combination experiments with very low doses of three genotoxic N-nitrosamines with similar organotropic carcinogenicity in rats. Carcinogenesis 8: 1635-1643. Bogovski, P. and Bogovski, S. (1981). Animal species in which N-nitroso compounds induce cancer. Int. J. Cancer 27: 471-474. da Costa Lopes, L.F., Albano, G.A.T., Laranja, L.M., Alves, L.F., Martins e Silva, G.P., Souza, Araujo, I.M., Nogueira-Neto, J.F., Felzenszwalb, I. and Kovary, K. (2000). Toxicological evaluation by in vitro and in vivo assays of an aqueous extract prepared from Echinodorus macroophyllus leaves. Toxicol. Lett. 116: 189-198. Lijinsky, W. (1992). Chemistry and Biology of N-Nitroso Compounds. Cambridge University Press, Cambridge, England. Maron, D.M. and Ames, B.N. (1983). Revised methods for the Salmonella mutagenicity test. Mutat. Res. 113: 173-215. McCann, J., Choi, E., Yamasaki, E. and Ames, B.N. (1975). Detection of carcinogens as mutagens in the Salmonella/microsome test: assay of 300 chemicals. Proc. Natl. Acad. Sci. USA 72: 5135-5139. Nakae, D., Kobayashi, Y., Akai, H. and Nabuaki, A. (1997). Involvement of 8-hydroxyguanine formation in the inhibition of rat liver carcinogenesis by low dose levels of N-nitrosodiethylamine. Cancer Res. 57: 1281-1287. Nakatsuru, Y., Matsukuma, S., Nemoto, N., Sugano, H., Sekiguchi, M. and Ishikawa, T. (1993). O6-Methyltransferase protects against nitrosamine-induced hepatocarcinogenesis. Proc. Natl. Acad. Sci. USA 90: 6468-6472. Pegg, E. (1983). Alkylating and subsequent repair of DNA after exposure to dimethylnitrosamine and related carcinogens. Rev. Biochem. Tox. 5: 88-123. Pegg, A.E. and Byers, T.L. (1992). Repair of DNA containing O6-alkylguanine. FASEB J. 6: 2302-2310. Ribeiro Pinto, L.F., Moraes, E., Albano, R.M., Silva, M.C., Godoy, W., Glisovic, T. and Lang, M.A. (2001). Rat oesophageal cytochrome P450 (CYP) monooxygenase system: comparison to the liver and relevance in N-nitrosodiethylamine carcinogenesis. Carcinogenesis 22: 1877-1883. Sander, J. (1967). Kann nitrit in der menschlichen nahrung ursache einer kerbsentstehung durch nitrosaminbildung sein? Arch. Hyg. Bakteriol. 151: 22-28. Tchou, T. and Grollman, A.P. (1993). Repair of DNA containing the oxidatively damaged base, 8-oxoguanine. Mutat. Res. 311: 277-287. Tricker, A.R. and Preussmann, R. (1991). Carcinogenic N-nitrosamines in the diet: occurrence, formation, mechanisms and carcinogenic potential. Mutat. Res. 259: 277-289. Vargas, V.M.F., Motta, V.E.P. and Henriques, J.A.P. (1993). Mutagenic activity detected by the Ames test in river water under the influence of petrochemical industries. Mutat. Res. 319: 31-45. Vargas, V.M.F., Guidobono, R.R., Jordão, C. and Henriques, J.A.P. (1995). Use of two short term tests to evaluate the genotoxicity of river water treated with different concentration/extraction procedures. Mutat. Res. 343: 31-52. Yoo, J.S.H., Ishizaki, H. and Yang, C.S. (1990). Roles of cytochrome P450 2E1 in the dealkylation and nitrosation of N-nitrosodimethylamine and N-nitrosodiethylamine in rat liver microsomes. Carcinogenesis 11: 2239-2243. Zeiger, E., Anderson, B., Haworth, S., Lawlor, T., Mortelmans, K. and Speck, W. (1987). Salmonella mutagenicity tests, III. Results from the testing of 255 chemicals. Environ. Mutagen. 9: 9-109. |
|