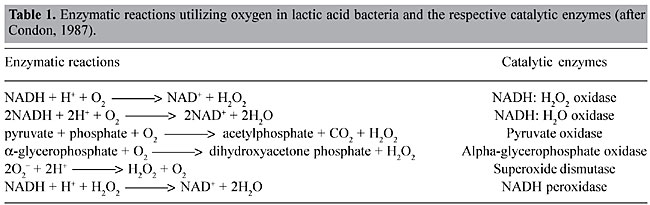
ABSTRACT. Lactococcus lactis, the most extensively characterized lactic acid bacterium, is a mesophilic- and microaerophilic-fermenting microorganism widely used for the production of fermented food products. During industrial processes, L. lactis is often exposed to multiple environmental stresses (low and high temperature, low pH, high osmotic pressure, nutrient starvation and oxidation) that can cause loss or reduction of bacterial viability, reproducibility, as well as organoleptic and/or fermentative qualities. Among these stress factors, oxidation can be considered one of the most deleterious to the cell, causing cellular damage at both molecular and metabolic levels. During the last two decades, considerable efforts have been made to improve our knowledge of oxidative stress in L. lactis. Many genes involved with both oxidative stress resistance and control mechanisms have been identified; functionally they seem to overlap. The finding of new genes, and a better understanding of the molecular mechanisms of stress resistance in L. lactis and other lactic acid baterium, will lead to the construction and isolation of stress-resistant strains. Such strains could be exploited for both traditional and probiotic uses. Key words: Lactococcus lactis, Oxygen, Stress INTRODUCTION Lactic acid bacteria (LAB) comprise a group of Gram-positive and facultative anaerobic microorganisms able to convert carbohydrates into lactic acid via homo- or heterofermentative metabolism (Stackebrandt and Teuber, 1988; Thompson, 1988). Due to this latter feature, some food-grade LAB species (Lactobacillus sp., Leuconostoc sp., Pediococcus sp., Streptococcus sp., and Lactococcus sp.) are widely used in the production of fermented food (vegetables, milk and meat), where these bacteria are responsible for both preservation and sensory characteristics, such as color, flavor and texture. Moreover, probiotic properties are attributed to some species of lactobacilli (Seegers, 2002). Lactococcus lactis is a mesophilic- (optimal growth temperature around 30°C) and microaerophilic-fermenting LAB (Duwat et al., 2000) with a relatively small genome (2.5 Mbp; Bolotin et al., 2001). This bacteria is commonly found in nature, on plant and animal surfaces and is widely used in the dairy industry for the production of fermented products, like cheese and buttermilk. Since over 107 tons of cheeses are annually produced worldwide, L. lactis is of great economic importance (Henriksen et al., 1999). Until the late 1970s, L. lactis ssp. lactis and L. lactis ssp. cremoris were empirically used by the dairy industry (Davidson et al., 1996; Henriksen et al., 1999). Thereafter, impelled by the advent of recombinant DNA technology, our knowledge about many biological aspects (e.g., microbiology, physiology, biochemistry, and mainly genetics) of these bacteria, and of other LAB, has greatly improved. Since then, the use of genetic tools has enabled the construction of many genetically modified strains that exhibit better controlled and/or improved growth and acidification, bacteriophage resistance and improved proteolytic properties (de Vos, 1999). Today, L. lactis is the most extensively characterized LAB (Bolotin et al., 2001), and it has been used to produce heterologous proteins of high biotechnological and medical interest, such as enzymes and antigens; it also has potential as a live vaccine (Langella and Le Loir, 1999). However, despite all the available data, the loss of some bacterial phenotypes, affecting viability, reproducibility, organoleptic, and/or fermentative capabilities, remains difficult to circumvent. In most cases, these problems are caused by the exposure of LAB to a range of stressful environmental conditions in both natural and/or industrial habitats. Industrial processing exposes L. lactis to multiple stress conditions, such as low and high temperature, low pH, high osmotic pressure, nutrient starvation, and oxidation. During the last two decades, several research groups have intensively studied stress responses in L. lactis. We describe general and specific aspects of oxidative stress in L. lactis and current insights related to the construction and isolation of both recombinant and non-recombinant L. lactis oxidative stress-resistant strains. OXIDATIVE STRESS Bacterial stress can be defined as a physiological perturbation, caused by environmental modifications (physical, chemical and/or nutritional) that can have many consequences for the bacteria, such as retarded growth and cell death (Farr and Kogoma, 1991; Fridovich, 1998; Duwat, 1999). Oxidative stress can cause several types of damage to the bacterial cell, including: i) metabolic pathway disruptions, ii) spontaneous mutations, and iii) bacteriostatic and bactericidal effects (Berlett and Stadtman, 1997; Fridovich, 1998). Oxygen by itself is unable to cause any damage to the cell; however, during the cellular processes (metabolic pathway), O2 is partially reduced to water, leading to the formation of reactive O2 species, which are the superoxide anion radical (O2-), the hydroxyl radical (OH·), and hydrogen peroxide (H2O2). These intermediates have a high oxidizing potential and thus are responsible for cellular oxygen toxicity (Farr and Kogoma, 1991; Fridovich, 1998; Storz and Imlay, 1999). OXIDATIVE STRESS IN LACTOCOCCUS LACTIS Described as a microaerophilic-fermenting LAB, L. lactis can, under certain conditions, tolerate and even use O2 (Condon, 1987; Duwat et al., 2001). Nevertheless, O2 consumption results in an altered redox state, and consequently greater NADH oxidase activity. Sugar fermentation is shifted towards mixed fermentation (Figure 1); hence, O2 participates in the oxidoreduction steps of NADH to NAD+. Through the action of NADH oxidases, H2O2 is formed (Table 1; Condon, 1987). The O2- can be generated through the following reaction: NADH + 2O2 ® NAD+ + H+ + 2O2-; which occurs because the flavin group of NADH oxidase carries out single-electron transfer, as well as the transfer of two or four electrons (Thomas and Pera, 1983; Imlay and Fridovich, 1991). The OH· can be formed during the Fenton reaction (H2O2 + Fe2+ + H+ ® OH· + H2O + Fe3+) (Duwat et al., 1995) or by spontaneous reactions (O2- + H2O2 ® OH- + OH· + O2) (Condon, 1987). Oxidative stress damages at the molecular level At the molecular level, O2-, OH· and H2O2 can react with cellular targets, such as proteins and nucleic acids. O2- has a moderate oxidizing potential and can attack compounds such as polyphenols, ascorbate and catecholamines (Farr and Kogoma, 1991; Fridovich, 1998). H2O2 can directly oxidize protein cysteinyl residues, thus inactivating enzymes (Storz and Imlay, 1999). It can also react with cations, such as Fe2+ and Cu+, and give rise to OH·, through the Fenton reaction (Farr and Kogoma, 1991; Duwat et al., 1995; Fridovich, 1998). OH· is a strong oxidant agent that can attack most organic compounds and can cause strand breaks and a wide spectrum of base modifications in DNA (Czapski, 1984; Farr and Kogoma, 1991; Fridovich, 1998). Another type of damage is the peroxidation of membrane lipids and membrane protein alterations, affecting cell permeability and osmoregulation (Harley et al., 1978; Kong and Davison, 1980). Oxidative stress damages at the metabolic level Many effects of O2 are also observed at the metabolic level. Under anaerobic conditions, L. lactis has a fermentative metabolism that enables the transformation of various types of carbohydrates into lactic acid (Figure 1) (Holt et al., 1994; Lopez de Felipe et al., 1997, 1998). In this case, 2 NADH molecules, generated from the oxidation of glyceraldehyde-3-phosphate, are reoxidized to favor the reduction of pyruvate to lactic acid by the action of lactate dehydrogenase (LDH). However, the NADH/NAD+ ratio plays a determinant role in the shift control from homolactic acid to mixed-acid fermentation in L. lactis (Garrigues et al., 1997). Under aerobic conditions, increased expression and activities of NADH oxidase and NADH peroxidase (Table 1) compete with LDH for NADH molecules (Murphy and Condon, 1984). Consequently, lactic acid production is reduced and glycolytic flux is shifted towards the production of acetate, ethanol, acetoin, diacetyl and CO2 (mixed-fermentation) by the action of pyruvate dehydrogenase (PDH), pyruvate-formate lyase and a-acetolactate synthase (Figure 1). These changes also lead to the formation of H2O2, which causes a reduction of the growth rate of L. lactis, and even its death. Concentrations of around 0.2 mM H2O2 inhibit the growth of this bacterium by 50% and concentrations >1.15 mM H2O2 can compromise cell viability (Anders et al., 1970; Duwat et al., 1999). Although NADH peroxidase (Table 1) contributes to cellular H2O2 detoxification, its activity is generally low (10 to 30 times lower than that of NADH oxidase) and endogenous or exogenous H2O2 overcomes the cell’s capability to deal with this oxygen species (Anders et al., 1970; Condon, 1987). 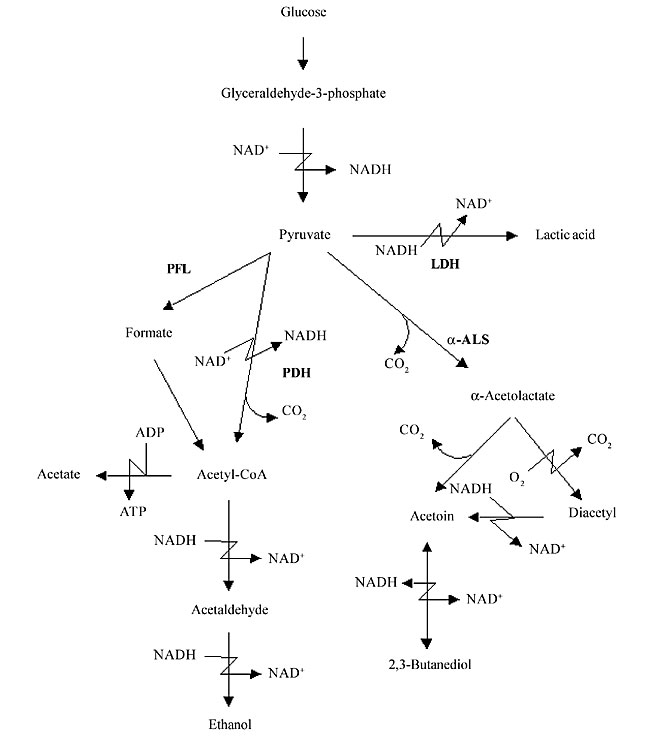
Figure 1. Schematic pathway of glucose metabolism in Lactococcus lactis. Intermediate and final glucose metabolism products are indicated by arrows. Catalytic enzymes are abbreviated in bold (LDH: lactate dehydrogenase; PDH: pyruvate dehydrogenase; PFL: pyruvate-formate lyase; a-ALS: a-acetolactate synthase).
OXIDATIVE STRESS RESISTANCE MECHANISMS IN LACTOCOCCUS LACTIS How does L. lactis overcome damage caused by active oxygen species? Experimental data and genomic analyses indicate that L. lactis, similar to Escherichia coli and Bacillus subtilis, is equipped with general and specific stress response mechanisms (Duwat et al., 1999). Several genes (see below) that participate in these mechanisms have been identified, and the respective encoded proteins have been shown to contribute to oxidative stress resistance. Moreover, induction of some of these genes is growth phase-dependent (exponential or stationary), and their products confer multi-stress resistance (Farr and Kogoma, 1991; Kjelleberg et al., 1993; Walker, 1996; Duwat et al., 2000). NADH oxidase/NADH peroxidase The most common oxidative stress resistance mechanism found in L. lactis, as well as in other LAB, is performed by a coupled NADH oxidase/NADH peroxidase system (Table 1; Condon, 1987). Initially, intracellular O2 can be eliminated by NADH oxidase activity, which utilizes O2 to oxidize NADH into NAD+, thereby giving rise to H2O2. Furthermore, NADH peroxidase reduces H2O2, generating H2O. However, NADH peroxidase activity is low, and considering that L. lactis does not possess any catalase activity, cellular H2O2 detoxification is inefficient. It was recently proposed that at high glycolytic fluxes, L. lactis ATCC19435 (L. lactis subsp. lactis, lacking both NADH peroxidase and superoxide dismutase (Sod) activities) metabolically produced pyruvate reacts nonenzymatically with H2O2 to form H2O and acetate (van Niel et al., 2002), thus providing the cell with an alternative mechanism of protection against H2O2. Superoxide dismutase A second common resistance mechanism is provided by the action of a Sod that removes O2- radicals (Table 1; Condon, 1987). In prokaryotes, the classification of these enzymes depends on the metal cofactor content (Cu-Zn, Fe or Mn). Most LAB have Sod activity. A unique manganese-containing Sod (MnSod) has been identified in L. lactis, during an analysis of acid stress-induced protein expression (Sanders et al., 1995). The growth rate of sodA mutants, under anaerobic conditions, is similar to that of the wild type strain, whereas under aerobic conditions the growth rate of this mutant strain is reduced (Sanders et al., 1995). Moreover, under anaerobic conditions, L. lactis sodA has a low initial expression, which is shifted to a high gene expression level under aerated conditions. An alternative mechanism to eliminate O2- radicals, which could compensate the low Sod activity, can be provided by high levels of intracellular glutathione (Fahey et al., 1978; Wiederholk and Steele, 1993). RecA Another oxidative stress resistance mechanism is supported by RecA activity. This protein is commonly found in bacteria, such as E. coli, where it plays a key role in the SOS response and homologous recombination (Miller and Kokjohn, 1990; Walker, 1996). Duwat et al. (1995) constructed a L. lactis recA mutant strain, and showed that RecA is involved in resistance to oxidative and thermal stresses. They observed that during exponential and stationary growth phases, recA mutants are highly sensitive to aeration, resulting in a reduction in growth rate and viability. They also postulated that in the absence of RecA activity, the OH· radical, generated during the Fenton reaction, is responsible for damage to DNA. To test this hypothesis, an iron chelator was added to aerated cultures of L. lactis recA mutants; bacterial doubling time was restored close to that of anaerobic conditions, leading to the observation that i) O2 radicals are responsible for oxygen toxicity and that ii) RecA plays an essential role in the repair of DNA damage caused by these compounds. They also studied the function of RecA under thermal stress. Under both oxidative and thermal stress, recA mutants have an elevated expression of HflB protein, which transcriptionally down-regulates heat-shock proteins, such as DnaK, GroEL and GrpE, and thus keeping these heat-shock proteins from being induced under elevated temperature conditions (above 37°C). Moreover, recA of L. lactis is co-transcribed with another DNA repair gene, formamidopyrimidine DNA glycosylase (fpg) (Duwat et al., 1992, 1995). In conclusion, activation of recA by oxidative stress can confer cross-protection against thermal stress. A single stimulus can activate various stress resistance mechanisms, conferring protection against various types of stress in L. lactis. Cytochrome d oxidase (cydA) Another way to cope with oxidative stress would be efficient O2 uptake and utilization, as in the case of bacteria able to respire. Despite its fermentative metabolism classification, there is evidence that L. lactis can undergo respiration in aerated culture media in the presence of heme (an iron-containing porphyrin molecule that forms the oxygen-binding portion of molecules such as hemoglobin) (Sijpesteijn, 1970). More recently, based on L. lactis IL1403 genome data, it was found that this bacterium possesses some of the genes needed for respiration, such as cydA, which encodes for cytochrome d oxidase (Bolotin et al., 1999). During respiration, CydA directly participates in the electron transfer chain, where O2 is the terminal electron acceptor, thus generating ATP. To test these possibilities and the respiration capacity of L. lactis, a cydA mutant strain was constructed (Duwat et al., 2001). Under aerated conditions, in the presence of heme, it was observed that i) L. lactis cydA expression is induced during the stationary growth phase; ii) this bacterium has an extended growth period and improved long-term survival, and iii) its metabolism is shifted from fermentation to respiration, as demonstrated by reduced lactic acid production and elevated acetate, acetoin and diacetyl formation. Based on this evidence, it is clear that L. lactis is able to respire, and probably this ability enables it to overcome oxidative stress injuries. Adaptation Another resistance mechanism in L. lactis is adaptation. It is known that in response to environmental changes, bacteria can trigger adaptive mechanisms that enable cells to tolerate harsh conditions (Condon, 1987; Woojin et al., 1999, 2002). Previously, it was observed that lactococci exposed to sublethal concentrations of H2O2 became able to grow in the presence of lethal concentrations (³1.15 mM) of H2O2 (Anders et al., 1970; Condon, 1987). More recently, it has been shown that adaptation to a specific stress, such as UV irradiation, starvation or high temperature, can lead to multi-stress resistance acquisition, probably due to the induction of genes that have functions in common in the resistance mechanisms against various types of stress (Duwat et al., 1995; Hartke et al., 1996, 1997; Duwat, 1999). CONTROL OF OXIDATIVE STRESS RESPONSES IN LACTOCOCCUS LACTIS Oxidative stress resistance mechanisms in L. lactis seem to be overlap, and thus cells under one specific stress condition can trigger different stress responses. Some studies have provided us with interesting clues concerning the control of oxidative stress responses in L. lactis (Duwat et al., 1999, 2000; Rallu et al., 2000; Scott et al., 2000; O’Connell-Motherway et al., 2000). Sigma factors Control at the transcriptional level, by sigma (s) factors would be an efficient way to modulate stress responses. However, whereas E. coli and B. subtilis have stress specific s-factors (Lange and Hengge-Aronis, 1994; Herman et al., 1995; Hecker et al., 1996), L. lactis only has three s-factors: RpoD, ComX and SigX (Araya et al., 1993; Bolotin et al., 1999, 2001). The vegetative s-factor, RpoD (Araya et al., 1993), allows the binding of RNA polymerase and the initiation of transcription at the so-called “vegetative promoters”, whereas ComX function is likely to induce competence genes (Bolotin et al., 1999, 2001). Although SigX, an extracytoplasmic function (Helmann, 2002) RNA polymerase s-factor in B. subtilis has been associated with heat stress resistance (Huang et al., 1997), no function for the lactococcal SigX has been proposed. Metabolite flux sensors The cAMP receptor protein, the fumarate and nitrate reduction regulator (FNR) and the FNR-like proteins (Flp) are members of a superfamily of structurally related transcriptional factors that could be involved in the control of stress responses (Green et al., 2001). Commonly found in a wide range of bacteria, these proteins possess structural properties that provide cells with versatile sensory (transducing environmental and metabolic signals) and DNA/RNA-regulatory mechanisms (DNA recognition motifs and RNA polymerase interaction) that trigger general and specific responses to particular physiological conditions, such as stress. In both E. coli (Jordan et al., 1997) and Lactobacillus casei (Gostick et al., 1998), FNR and Flp play a key role in oxidative stress by interaction with an oxygen-labile [4Fe-4S] cluster. L. lactis possesses two Flp encoding genes, flpA and flpB, located in two paralogous operons (orfX-orfY-flp), where the proximal genes appear to encode potential metal ion transport systems (Gostick et al., 1999). To further characterize the L. lactis Flp proteins, single and double mutants of flpA/B were constructed (Scott et al., 2000). No phenotypic effects were observed between the single mutant and the wild-type strain when submitted to oxidative stress conditions (H2O2). However, under the same conditions the double mutant was more affected, as evidenced by i) an increased lag phase, ii) an altered polypeptide profile, and iii) a lower intracellular zinc [Zn(II)] content. This indicates that at least one of the L. lactis Flp must be functional in order to be able to cope with oxidative stress. Based on these observations, it was suggested that FlpA/B may be involved in the control of Zn(II) uptake and stress resistance mechanisms. It was also postulated that L. lactis uses Zn(II) to protect protein thiol groups from oxidation and that intracellular levels of this compound may be functioning as a “metabolite flux sensor” (MFS), sensing and controlling general stress responses. An example of how MFS can control stress responses was given by the construction of L. lactis recA-derived mutant strains. The L. lactis recA strain is sensitive to both oxidative and thermal stresses. Therefore, in order to identify other mechanisms related to this sensitivity, recA-derived strains were isolated by random insertional mutagenesis (Duwat et al., 1999). Most mutated genes were found to encode proteins involved in metabolic pathways for the biosynthesis of guanosine phosphate (GP) (deoB, guaA and tktA) or in phosphate uptake (pstS and pstB). These mutations conferred resistance to multiple stresses and it was postulated that low intracellular levels of GP and/or phosphate are functioning as MFS, controlling general stress responses. This hypothesis was later confirmed by Duwat et al., 2000, who showed that depletion of GP in growth media of wild-type strains was able to induce general stress resistance; otherwise, the addition of guanine (over 15 µg/ml) to the growth medium from a guaA/recA-derivative mutant strain resulted in complete loss of the stress-resistant phenotype. The involvement of phosphate pools was also demonstrated with the pstB/recA-derivative mutant strain (Duwat et al., 2000). Similar results were obtained during an independent selection of acid-resistant mutants of L. lactis, which were also found to be resistant to oxidative stress caused by H2O2 (Rallu et al., 2000). Two-component regulatory systems Control of stress responses could also be achieved by cellular systems that sense and transmit environmental signals into the cell, thereby modulating gene expression and physiological changes. These systems have been described in both prokaryotic and eukaryotic organisms, and are known as “two-component regulatory systems” (Chang and Stewart, 1998). Basically, these systems are composed of a membrane-anchored sensor protein, usually a histidine protein kinase, and an intracellular response regulator. In L. lactis, six of these systems have been identified (O’Connell-Motherway et al., 1997) and three of these (systems B, D and F) are involved in specific susceptibility to acid, osmotic and oxidative stress (O’Connell-Motherway et al., 2000). In the latter (system F), the L. lactis mutant strain, obtained by insertional mutagenesis, has greater H2O2 sensitivity than the wild-type strain (O’Connell-Motherway et al., 2000). At least in this case, it seems that the control of stress responses are stress specific. In summary, many questions remain and it is possible that control of stress resistance mechanisms in L. lactis operates through pleiotropic effects or even through multiple regulatory mechanisms, some of which may be unique to this bacterium. PRESENT AND FUTURE DIRECTIONS Despite all the efforts that have been made to identify and improve our knowledge about oxidative stress resistance and its control mechanisms in L. lactis, a number of questions remain unanswered. Many aspects of general and specific stress in this bacterium differ from model microorganisms, such as E. coli and B. subtilis. Thus, it becomes an important challenge to isolate and construct new stress resistant strains that can help to answer these questions. Most studies concerning stress in L. lactis concern the production of stress-resistant mutant strains; depending on the method employed to obtain these strains, they can be classified as: i) “non-genetically modified mutant strains” (non-GMM) or ii) “genetically modified mutant strains” (GMM). In the former, stress resistant strains are acquired through screening in selective media, or are isolated from industrial sources or from their natural habitat. Nevertheless, these types of mutants are not very informative, due to difficulties in identifying possible genes and/or metabolic pathways involved in stress resistance. Alternatively, such genes or pathways can be characterized by the identification and analysis of stress-induced proteins in two-dimensional polyacrylamide gel electrophoresis (Giard et al., 2002). GMM strains can be constructed by site-directed mutagenesis (homologous recombination utilizing non-replicative plasmids; Leloup et al., 1997; O’Connell-Motherway et al., 2000) or by random insertional mutagenesis (transposition elements; Schafer et al., 1991; Maguin et al., 1992). One of the most powerful tools that has been developed is the thermo-sensitive plasmid pG+host:ISS1 (Maguin et al., 1996), which permits insertional gene inactivation and subsequent determination of its involvement in bacterial stress. In addition to basic knowledge of the L. lactis stress response, obtained through the construction of stress resistant strains, new information about oxidative and other types of stress has opened new possibilities for their practical application. Today, similar findings have been applied to the metabolic engineering of sugars and other molecules (Lopez de Felipe et al., 1997, 1998; Hugenholtz et al., 2002; Koebmann et al., 2002; Cocaign-Bousquet et al., 2002), in the development of new expression systems (Sanders et al., 1998; Madsen et al., 1999), and to develop new anti-oxidant strains of L. lactis that could become useful in medicine and industry. ACKNOWLEDGMENTS We are grateful to Elisabeth Azevedo, Herbert P. Schweizer, and Santuza Teixeira for discussion and text corrections. We are also grateful to Alexandra Gruss for scientific discussion and to our team members and colleagues (Brazilian and French) for their friendly support. We have a special thought for Patrick Duwat (deceased January 5, 2000) who initiated the work on L. lactis oxidative stress in the URLGA-INRA. Research supported by grants from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil) and COFECUB (Comité Français d’Evaluation de la Coopération Universitaire avec le Brésil). REFERENCES Anders, R.S., Hogg, D.M. and Jago, G.R. (1970). Formation of H2O2 by group N streptococci and its effect on their growth and metabolism. Appl. Microbiol. 19: 608-612. Araya, T., Ishibashi, N., Shimamura, S., Tanaka, K. and Takahashi, H. (1993). Genetic and molecular analysis of the rpoD gene from Lactococcus lactis. Biosci. Biotech. Biochem. 57: 88-92. Berlett, B.S. and Stadtman, E.R. (1997). Protein oxidation in aging, disease and oxidative stress. J. Biol. Chem. 272: 20313-20316. Bolotin, A., Mauger, S., Malarme, K., Ehrlich, S.D. and Sorokin, A. (1999). Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Van Leeuwenhoek 76: 27-76. Bolotin, A., Wincker, P., Mauger, S., Jaillon, O., Malarme, K., Weissenbach, J., Ehrlich, S.D. and Sorokin, A. (2001). The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp lactis IL1403. Genome Res. 11: 731-753. Chang, C. and Stewart, R.C. (1998). The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 117: 723-731. Cocaign-Bousquet, M., Even, S., Lindley, N.D. and Loubiere, P. (2002). Anaerobic sugar catabolism in Lactococcus lactis: genetic regulation and enzyme control over pathway flux. Appl. Microbiol. Biot. 60: 24-32. Condon, S. (1987). Responses of lactic acid bacteria to oxygen. FEMS Microbiol. Rev. 46: 269-280. Czapski, G. (1984). Reaction of OH. Methods Enzymol. 105: 209-215. Davidson, B.E., Kordias, N., Dobos, M. and Hillier, A.J. (1996). Genomic organization of lactic acid bacteria. Antonie Van Leeuwenhoek 70: 161-183. de Vos, W.M. (1999). Gene expression systems for lactic acid bacteria. Curr. Opin. Microbiol. 2: 289-295. Duwat, P. (1999). Stress response pathways in Lactococcus lactis. Recent Res. Devel. Microbiol. 3: 335-348. Duwat, P., Ehrlich, S.D. and Gruss, A. (1992). Use of degenerate primers for polymerase chain reaction cloning and sequencing of the Lactococcus lactis subsp. lactis recA gene. J. Bacteriol. 174: 5171-5175. Duwat, P., Ehrlich, S.D. and Gruss, A. (1995). The recA gene of Lactococcus lactis: characterization and involvement in oxidative and thermal stress. Mol. Microbiol. 17: 1121-1131. Duwat, P., Ehrlich, S.D. and Gruss, A. (1999). Effects of metabolic flux on stress response pathways in Lactococcus lactis. Mol. Microbiol. 31: 845-858. Duwat, P., Cesselin, B., Sourice, S. and Gruss, A. (2000). Lactococcus lactis, a bacterial model for stress responses and survival. Int. J. Food Microbiol. 55: 83-86. Duwat, P., Sourice, S., Cesselin, B., Lamberet, G., Vido, K., Gaudu, P., Le Loir, Y., Violet, F., Loubière, P. and Gruss, A. (2001). Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J. Bacteriol. 183: 4509-4516. Fahey, R.C., Brown, W.C., Adams, W.B. and Worsham, M.B. (1978). Occurrence of glutathione in bacteria. J. Bacteriol. 133: 1126-1129. Farr, S.B. and Kogoma, T. (1991). Oxidative stress responses in E. coli and S. typhimurium. Microbiol. Rev. 55: 561-585. Fridovich, I. (1998). Oxygen toxicity: a radical explanation. J. Exp. Biol. 201: 1203-1209. Garrigues, C., Loubiere, P., Lindley, N.D. and Cocaign-Bousquet, M. (1997). Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179: 5282-5287. Giard, J.C., Verneuil, N., Auffray, Y. and Hartke, A. (2002). Characterization of genes homologous to the general stress-inducible gene gls24 in Enterococcus faecalis and Lactococcus lactis. FEMS Microbiol. Lett. 206: 235-239. Gostick, D.O., Green, J., Irvine, A.S., Gasson, M.J. and Guest, J.R. (1998). A novel regulatory switch mediated by the FNR-like protein of Lactobacillus casei. Microbiology 144: 705-717. Gostick, D.O., Griffin, H.G., Shearman, C.A., Scott, C., Green, J., Gasson, M.J. and Guest, J.R. (1999). Two operons that encode FNR-like proteins in Lactococcus lactis. Mol. Microbiol. 31: 1523-1535. Green, J., Scott, C. and Guest, J.R. (2001). Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv. Microb. Physiol. 44: 1-34. Harley, J.B., Santangelo, G.M., Rasmussen, H. and Goldfine, H. (1978). Dependence of Escherichia coli hyperbaric oxygen toxicity on the lipid acyl chain composition. J. Bacteriol. 134: 808-820. Hartke, A., Bouche, S., Giard, J.C., Benachour, A., Boutibonnes, P. and Auffray, Y. (1996). The lactic acid stress response of Lactococcus lactis subsp. lactis. Curr. Microbiol. 33: 194-199. Hartke, A., Frere, J., Boutibonnes, P. and Auffray, Y. (1997). Differential induction of the chaperonin GroEL and the Co-chaperonin GroES by heat, acid, and UV-irradiation in Lactococcus lactis subsp. lactis. Curr. Microbiol. 34: 23-26. Hecker, M., Schumann, W. and Volker, U. (1996). Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19: 417-428. Helmann, J.D. (2002). The extracytoplasmic function (ECF) sigma factors. Adv. Microbiol. Physiol. 46: 47-110. Henriksen, C.M., Nilson, D., Hansen, S. and Johansen, E. (1999). Industrial applications of genetically modified microorganisms: gene technology at Chr. Hansen A/S. Intern. Dairy J. 9: 17-23. Herman, C., Lecat, S., D’Ari, R. and Bouloc, P. (1995). Regulation of the heat-shock response depends on divalent metal ions in an hflB mutant of Escherichia coli. Mol. Microbiol. 18: 247-255. Holt, J.G., Krieg, N.R., Sneath, P.H.A., Staley, J.T. and Williams, S.T. (1994). Gram-positive cocci. In: Bergey’s Manual of Determinative Microbiology (Holt, J.G., ed.). Williams & Wilkins, Baltimore, MD, USA, pp. 527-558. Huang, X., Decatur, A., Sorokin, A. and Helmann, J.D. (1997). The Bacillus subtilis sigma(X) protein is an extracytoplasmic function sigma factor contributing to survival at high temperature. J. Bacteriol. 179: 2915-2921. Hugenholtz, J., Sybesma, W., Groot, M.N., Wisselink, W., Ladero, V., Burgess, K., van Sinderen, D., Piard, J.C., Eggink, G., Smid, E.J., Savoy, G., Sesma, F., Jansen, T., Hols, P. and Kleerebezem, M. (2002). Metabolic engineering of lactic acid bacteria for the production of nutraceuticals. Antonie Van Leeuwenhoek 82: 217-235. Imlay, J.A. and Fridovich, I. (1991). Assay of metabolic superoxide production in Escherichia coli. J. Biol. Chem. 266: 6957-6965. Jordan, P.A., Thomson, A.J., Ralph, E.T., Guest, J.R. and Green, J. (1997). FNR is a direct oxygen sensor having a biphasic response curve. FEBS Lett. 416: 349-352. Kjelleberg, S., Albertson, N., Flardh, K., Holmquist, L., Jouper-Jaan, A., Marouga, R., Ostling, J., Svenblad, B. and Weichart, D. (1993). How do non-differentiating bacteria adapt to starvation? Antonie Van Leeuwenhoek 63: 333-341. Koebmann, B.J., Andersen, H.W., Solem, C. and Jensen, P.R. (2002). Experimental determination of control of glycolysis in Lactococcus lactis. Antonie Van Leeuwenhoek 82: 237-248. Kong, S. and Davison, A.J. (1980). The role of interactions between O2, H2O2, ·OH,e- and O2- in free radical damage to biological systems. Arch. Biochem. Biophys. 204: 18-29. Lange, R. and Hengge-Aronis, R. (1994). The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8: 1600-1612. Langella, P. and Le Loir, Y. (1999). Heterologous protein secretion in Lactococcus lactis: a novel antigen delivery system. Braz. J. Med. Biol. Res. 32: 191-198. Leloup, L., Ehrlich, S.D., Zagorec, M. and Morel-Deville, F. (1997). Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 63: 2117-2123. Lopez de Felipe, F., Starrenburg, M.J.C. and Hugenholtz, J. (1997). The role of NADH-oxidation in acetoin and diacetyl production from glucose in Lactococcus lactis subsp. lactis MG1363. FEMS Microbiol. Lett. 156: 15-19. Lopez de Felipe, F., Kleerebezem, M., de Vos, W.M. and Hugenholtz, J. (1998). Cofactor engineering: a novel approach to metabolic engineering in Lactococcus lactis by controlled expression of NADH oxidase. J. Bacteriol. 180: 3804-3808. Madsen, S.M., Arnau, J., Vrang, A., Givskov, M. and Israelsen, H. (1999). Molecular characterization of the pH-inducible and growth phase-dependent promoter P170 of Lactococcus lactis. Mol. Microbiol. 32: 75-87. Maguin, E., Duwat, P., Hege, T., Ehrlich, S.D. and Gruss, A. (1992). New thermosensitive plasmid for Gram-positive bacteria. J. Bacteriol. 174: 5633-5638. Maguin, E., Prevost, H., Ehrlich, S.D. and Gruss, A. (1996). Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178: 931-935. Miller, R.V. and Kokjohn, T.A. (1990). General microbiology of recA: environmental and evolutionary significance. Ann. Rev. Microbiol. 44: 365-394. Murphy, M.G. and Condon, S. (1984). Correlation of oxygen utilization and hydrogen peroxide accumulation with oxygen induced enzymes in Lactobacillus plantarum cultures. Arch. Microbiol. 138: 44-48. O’Connell-Motherway, M., Fitzgerald, G.F. and van Sinderen, D. (1997). Cloning and sequence analysis of putative histidine protein kinases isolated from Lactococcus lactis MG1363. Appl. Environ. Microbiol. 63: 2454-2459. O’Connell-Motherway, M., van Sinderen, D., Morel-Deville, F., Fitzgerald, G.F., Ehrlich, S.D. and Morel, P. (2000). Six putative two-component regulatory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology 146: 935-947. Rallu F., Gruss, A., Ehrlich, S.D. and Magnin, E. (2000). Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35: 517-528. Sanders, J.W., Leenhouts, K.J., Haandrikman, A.J., Venema, G. and Kok, J. (1995). Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J. Bacteriol. 177: 5254-5260. Sanders, J.W., Venema, G., Kok, J. and Leenhouts, K. (1998). Identification of a sodium chloride-regulated promoter in Lactococcus lactis by single-copy chromosomal fusion with a reporter gene. Mol. Gen. Genet. 257: 681-685. Schafer, A., Jahns, A., Geis, A. and Teuber, M. (1991). Distribution of the IS elements ISS1 and IS904 in lactococci. FEMS Microbiol. Lett. 64: 311-317. Scott, C., Rawsthorne, H., Upadhyay, M., Shearman, C.A., Gasson, M.J., Guest, J.R. and Green, J. (2000). Zinc uptake, oxidative stress and the FNR-like proteins of Lactococcus lactis. FEMS Microbiol. Lett. 192: 85-89. Seegers, J.F. (2002). Lactobacilli as live vaccine delivery vectors: progress and prospects. Trends Biotechnol. 20: 508-515. Sijpesteijn, A.K. (1970). Induction of cytochrome formation and stimulation of oxidative dissimilation by hemin in Streptococcus lactis and Leuconostoc mesenteroides. Antonie Van Leeuwenhoek 36: 335-348. Stackebrandt, E. and Teuber, M. (1988). Molecular taxonomy and phylogenetic position of lactic acid bacteria. Biochimie 70: 317-324. Storz, G. and Imlay, J.A. (1999). Oxidative stress. Curr. Opin. Microbiol. 2: 188-194. Thomas, E.L. and Pera, K.A. (1983). Oxygen metabolism of Streptococcus mutans: uptake of oxygen and release of superoxide and hydrogen peroxide. J. Bacteriol. 154: 1236-1244. Thompson, J. (1988). Lactic acid bacteria: model systems for in vivo studies of sugar transport and metabolism in gram-positive organisms. Biochimie 70: 325-336. van Niel, E.W., Hofvendahl, K. and Hahn-Hagerdal, B. (2002). Formation and conversion of oxygen metabolites by Lactococcus lactis subsp. lactis ATCC 19435 under different growth conditions. Appl. Environ. Microbiol. 68: 4350-4356. Walker, G.C. (1996). The SOS response of Escherichia coli. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology (Neidhardt, F.C., ed.). ASM, Washington, DC, USA, pp. 1400-1416. Wiederholk, K.M. and Steele, J.L. (1993). Glutathione accumulation in lactococci. J. Dairy Sci. 77: 1183-1188. Woojin, S.K., Ren, J. and Dunn, N.W. (1999). Differentiation of Lactococcus lactis subspecies lactis and subspecies cremoris strains by their adaptive response to stresses. FEMS Microbiol. Lett. 171: 57-65. Woojin, S.K., Park, J.H., Tandianus, J.E., Ren, J., Su, P. and Dunn, N.W. (2002). A distinct physiological state of Lactococcus lactis cells that confers survival against a direct and prolonged exposure to severe stresses. FEMS Microbiol. Lett. 212: 203-208. |
|