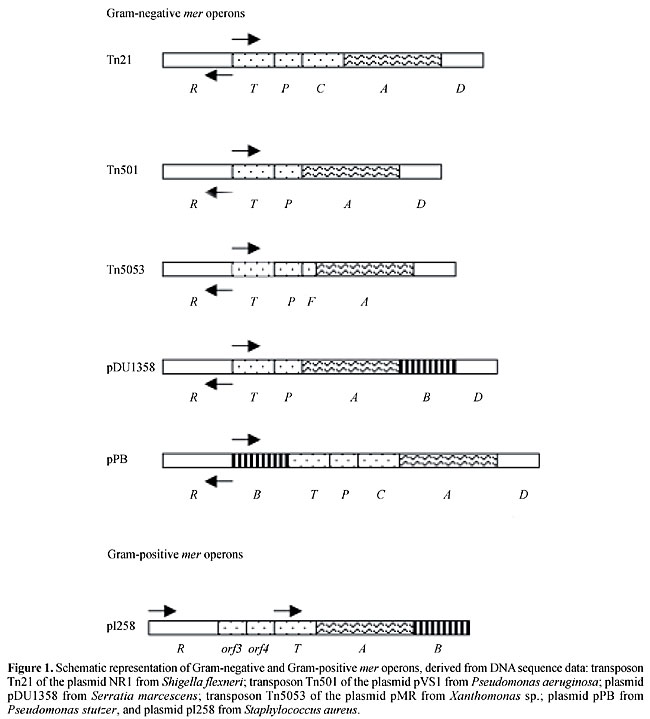
ABSTRACT. Mercury is present in the environment as a result of natural processes and from anthropogenic sources. The amount of mercury mobilized and released into the biosphere has increased since the beginning of the industrial age. Generally, mercury accumulates upwards through aquatic food chains, so that organisms at higher trophic levels have higher mercury concentrations. Some bacteria are able to resist heavy metal contamination through chemical transformation by reduction, oxidation, methylation and demethylation. One of the best understood biological systems for detoxifying organometallic or inorganic compounds involves the mer operon. The mer determinants, RTPCDAB, in these bacteria are often located in plasmids or transposons and can also be found in chromosomes. There are two classes of mercury resistance: narrow-spectrum specifies resistance to inorganic mercury, while broad-spectrum includes resistance to organomercurials, encoded by the gene merB. The regulatory gene merR is transcribed from a promoter that is divergently oriented from the promoter for the other mer genes. MerR regulates the expression of the structural genes of the operon in both a positive and a negative fashion. Resistance is due to Hg2+ being taken up into the cell and delivered to the NADPH-dependent flavoenzyme mercuric reductase, which catalyzes the two-electron reduction of Hg2+ to volatile, low-toxicity Hg0. The potential for bioremediation applications of the microbial mer operon has been long recognized; consequently, Escherichia coli and other wild and genetically engineered organisms for the bioremediation of Hg2+-contaminated environments have been assayed by several laboratories. Key words: Operon mer, Bacterial mercury resistance, Bioremediation INTRODUCTION Mercury, the 6th most toxic in a universe of 6 million substances, exists naturally in small amounts in the environment, being the 16th most rare element on Earth. However, its levels have risen due to environmental contamination from human activities, such as burning coal and petroleum products, use of mercurial fungicides in paper making and agriculture and mercury catalysts in industry, with a consequent release of mercury into the air and water and on the land. These activities can increase local mercury levels several thousand-fold above background (Tuovinen, 1984). In Brazil, huge amounts of mercury are used at prospecting sites for amalgam formation in gold extraction. An average of 1.32 kg of mercury is used for each kilogram of gold produced (Lacerda and Solomons, 1991). As a consequence, metallic mercury is introduced into the environment, representing one of the major sources of aggression against man and the environment. Its use in seed and bulb dressings directed against bacteria and fungi on fruit trees has introduced much of the mercury that contaminates agricultural land. Therefore, environmental pollution is an increasing problem both for developing and developed countries. Minamata disease, discovered in 1956 around Minamata Bay, Japan, is the first instance on record of severe methylmercury poisoning, having affected thousands of people, 887 of whom were killed (Daher, 1999). It resulted from the consumption, mainly by fishermen and their families, of large amounts of fish and shellfish contaminated with methylmercury, resulting from the transformation of the HgCl2 discharged from a chemical plant (Chisso Co. Ltd.). Methylmercury is a neurological poison primarily affecting the central nervous system, liver and kidney. When ingested, almost all of the methylmercury is absorbed. Its half-life is about 44 days. Most methylmercury is converted and excreted into the feces and urine (Abelsohn et al., 2002). The other chemical forms of mercury, vapor and inorganic mercury, accumulate in the brain (Hg0) and kidney (Hg2+). The kidney is the main target organ for inorganic mercury. The typical symptoms of Minamata disease include neurological disorders, such as sensory disorders, cerebellar ataxia, constriction of the visual field, auditory disturbances, tremoring of the visual field, and disequilibrium (Langford and Ferner, 1999). Furthermore, many of the affected individuals in Minamata were congenitally affected by methylmercury. Their mothers had only mild or no manifestation of poisoning (Harada, 1978). This fact demonstrates the much higher vulnerability of fetuses than adults and shows that methylmercury easily passes through the placenta and affects the fetus (Nishigaki and Harada, 1975). MERCURY CYCLE IN THE ENVIRONMENT The environmental mercury cycle is mediated by both geological and biological processes. Mercury vapor (metallic mercury) emitted from both natural and anthropogenic sources is globally distributed in the atmosphere. The major form of mercury in the atmosphere is vapor mercury (Hg0), which is volatile and is oxidized to mercuric ion (Hg2+) as a result of its interaction with ozone in the presence of water (Munthe and McElroy, 1992; DeMagalhaes and Tubino, 1995; Pleijel and Munthe, 1995). Most of the mercury entering aquatic environments is Hg2+. Inorganic mercury, present in water and sediments, is subject to bacterial conversion to methylmercury compounds that are bioaccumulated in the aquatic food chain. Organomercury compounds are translocated rapidly through the food chain, with tragic consequences. Predatory organisms at the top of the food chain generally have higher mercury concentrations, found as organic forms of methylmercury. The major chemical forms of mercury to which humans are exposed are mercury vapor, Hg0, and methylmercury compounds, which are highly toxic to all living organisms. The toxicity of inorganic and organic mercury compounds is due to their strong affinity for sulfur-containing organic compounds, such as enzymes and other proteins. For this reason these compounds are extremely toxic to biological systems. However, bacteria, fungi and plants have evolved mechanisms of resistance to several of these different chemical forms. The bacteria play a major role in the global cycling of mercury in the natural environment. Bacterial resistance to mercury and their role in mercury cycling have been extensively studied (Osborn et al., 1997). This mini-review focuses predominantly on mercury resistance mer operons. BIOCHEMICAL BASIS OF BACTERIAL MERCURY RESISTANCE As a response to toxic mercury compounds globally distributed by geological and anthropogenic activities, microorganisms have developed a surprising array of resistance systems to overcome the poisonous environment. An extensively studied resistance system, based on clustered genes in an operon (mer operon), allows bacteria to detoxify Hg2+ into volatile metallic mercury by enzymatic reduction (Komura and Izaki, 1971; Summers, 1986; Misra, 1992; Silver, 1996; Osborn et al., 1997). Mercury-resistance determinants have been found in a wide range of Gram-negative and Gram-positive bacteria isolated from different environments. They vary in the number and identity of genes involved and are encoded by mer operons, usually located on plasmids (Summers and Silver, 1972; Brown et al., 1986; Griffin et al., 1987; Radstrom et al., 1994) and chromosomes (Wang et al., 1987; Inoue et al., 1991); they are often components of transposons (Misra et al., 1984; Kholodii et al., 1993) and integrons (Liebert et al., 1999). Two main mer determinant types have been described: narrow-spectrum mer determinants confer resistance to inorganic mercury salts only, whereas broad-spectrum mer determinants confer resistance to organomercurials such as methylmercury and phenylmercury, as well as to inorganic mercury salts (Misra, 1992; Silver and Phung, 1996; Bogdanova et al., 1998). The biochemical basis of resistance to inorganic mercury compounds such as HgCl2 appears to be quite similar in several different species. It involves the reduction of Hg2+ to volatile Hg0 by an inducible enzyme, mercuric reductase. This enzyme has been characterized in plasmid-carrying strains of Pseudomonas, Escherichia coli and Staphylococcus aureus (Summers and Silver, 1978; Bhriain and Foster, 1996; Silver and Phung, 1996; Osborn et al., 1997). This reductase is a flavoprotein, which catalyzes the NADPH-dependent reduction of Hg2+ to Hg0. Since mercury has such a high vapor pressure, it volatilizes and the bacterial environment is left mercury free. This mercuric reductase is found intracellularly and is inducible by subinhibitory concentrations of mercuric ions and a variety of organomercurial substances (Furukawa and Tonomura, 1972; Summers, 1972; Schottel, 1978). Based on a comparison with other bacterial periplasmic binding, protein-dependent transport systems, it has been proposed that Hg2+ diffuses across the outer membrane (Brown, 1985). Mercuric ions are transported outside the cell by a series of transporter proteins. This mechanism involves the binding of Hg2+ by a pair of cysteine residues on the MerP protein located in the periplasm. Hg2+ is then transferred to a pair of cysteine residues on MerT, a cytoplasmic membrane protein, and finally to a cysteine pair at the active site of MerA (mercuric reductase) (Hamlett et al., 1992). Next, Hg2+ is reduced to Hg0 in an NADPH-dependent reaction. The non-toxic Hg0 is then released into the cytoplasm and volatilizes from the cell. The biochemical mechanism for broad-spectrum resistance to organomercurials involves, in addition to mercuric reductase, another inducible, soluble enzyme: organomercurial lyase. This enzyme cleaves the organometallic linkage to yield Hg2+, and then the reductase uses NADPH to reduce the elemental mercury form, which volatilizes from the cell (Schottel, 1978). STRUCTURE OF THE MER OPERON The mer operons vary in structure and are constituted by genes that encode the functional proteins for regulation (merR), transport (merT, merP and/or merC, merF) and reduction (merA) (Figure 1). In some cases, known as broad-spectrum mercury resistance, additional merB genes are required to confer resistance to many organomercurials, such as methylmercury and phenylmercury, by hydrolyzing the C-Hg bond before Hg2+ reduction. In general, the additional merB genes are found downstream of the merA gene in the mer operon (Osborn et al., 1997).
Most mer operons contain a regulatory gene, mer R, which is transcribed separately and divergently from the structural mer genes. However, in Gram-positive bacteria the merR genes of pI258 from Staphylococcus aureus and RC 607 from Bacillus sp. are transcribed in the same direction as the structural genes (Laddaga et al., 1987; Wang et al., 1989). MerR, the metalloregulatory protein, binds the promoter-operator region, where it both positively and negatively regulates the expression of the divergently transcribed structural genes, and also negatively regulates its own expression. MerR protein activates transcription of the operon in the presence of inducing concentrations of Hg2+. It represses transcription of the structural genes from the mer operon (merTPCFAD) in the absence of Hg2+, and activates transcription in the presence of Hg2+. The most distal promoter gene, merD, which is co-transcribed with the structural genes, down-regulates the mer operon. MerD, a secondary regulatory protein, also binds the same operator-promoter region as MerR, although very weakly (Nucifora et al., 1990; Mukhopadhyay et al., 1991). A number of structural genes are found downstream of the operator/promoter site; the proteins they code for are involved in mercuric ion transport. All the mer operons have merT and merP, however, some operons, such as transposon Tn21, have merC (the first example found with the merC gene). The additional merC gene is located between merP and merA. However, it seems not to be essential for Hg2+ resistance since it is absent from Tn501, which confers identical Hg2+ resistance levels (Bhriain and Foster, 1986; Summers, 1986). Both merT and merP are required for full expression of Hg2+ resistance, but loss of merP is less deleterious than loss of merT. In contrast, mutating merC had no effect on Hg2+ resistance, though it decreased the level of expression. Recently, one more mer gene implicated in mercuric transport, merF, was found in plasmid pMER327/419 of Pseudomonas fluorescens between merP and merA (Wilson et al., 2000). The merA gene, determining mercuric reductase, and merB, if present, encoding the enzyme organomercurial lyase, are immediately followed by genes encoding transport function. However, as observed in Pseudomonas stutzeri, the merB gene is found between merR and merT, together with an extra operator-promoter region (Weiss et al., 1977; Walsh et al., 1988; Reniero et al., 1995). The other genes encoding organomercury resistance have been identified and designated merG and merE, located between merA and merB on the broad-spectrum mer operon (Huang et al., 1999; Kiyono and Pan-Hou, 1999). Furthermore, merB seldom occurs in Gram-negative bacteria (Laddaga et al., 1987; Wang et al., 1989; Sedlmeier and Altenbuchner, 1992; Bogdanova et al., 1998). Various mercury detoxification mechanisms, without mercury-reducing activity, have been reported, such as reduced uptake of mercuric ions due to reduction in cellular permeability to Hg2+ ions (Pan-Hou et al., 1981), demethylation of methylmercury by Clostridium cochlearium T-2P, which involves the decomposition and inactivation of inorganic mercury with hydrogen sulfide (H2S) (Pan-Hou and Imura, 1981), mercury methylation by certain bacteria that use methylation as a resistance/detoxification mechanism (Trevors, 1986) and sequestration of methylmercury (Silver and Misra, 1984). MERCURY AND ANTIBIOTIC RESISTANCE Mercury pollution can contribute to increased antibiotic resistance (McArthur and Tuckfield, 2000). The combined expression of antibiotic resistance and mercury may be caused by selection, as a consequence of the mercury present in an environment (Sant’ana et al., 1989). Mercury resistance operons, which are often found in conjugative plasmids and transposons, provide a suitable model system for the study of horizontal gene transfer in natural populations of bacteria. Bacterial plasmid resistance systems (mer gene) for mercurials and organomercurials are the best understood of such systems at the biochemical and molecular genetic levels (Kalyaeva et al., 1988; Silver, 1994). BIOTECHNOLOGICAL APPLICATIONS OF MER GENES TO MERCURY DECONTAMINATION AND RECOVERY Industrial use of mercury led to pollution of the environment. Consequently, mercury removal is a challenge for environmental management. Common processes to remove mercury from contaminated sources, based on adsorption with ion-exchange resins or biosorbents, have been found to be sensitive to environment conditions (Ritter and Bibbler, 1992; Chang and Hong, 1994). Biological processes have been employed in bioremediation, including metal recovery, and are potentially low cost. The use of bacteria for removing metal from contaminated environments is a promising technology. However, passive adsorption and immobilization treatments produce a large volume of mercury-loaded biomass, the disposal of which is problematic. Microorganisms in contaminated environments have developed resistance to mercury and are playing a major role in natural decontamination (Cursino et al., 1999). The bacterial plasmid/transposon resistance systems for mercurials and organomercurials (mer systems) are the best understood at the biochemical and molecular genetic levels (Silver, 1994), and are of great interest since they represent a natural strategy for the detoxification of mercury-contaminated environments. The potential of the microbial mer operon, which functions by active enzymatic reduction of mercury ions to water-insoluble metallic mercury, has been recognized for a long time, because of its high levels of efficacy and specificity. Inside the cell, Hg2+ is reduced to metallic mercury (Hg0), which passively diffuses out of the cell and its environment (with no energy expenditure) (Saouter et al., 1994; Silver, 1996; Silver and Phung, 1996; von Canstein et al., 1999; Chen et al., 1999; Nies, 1999). Therefore, the bacterial biomass acts continuously as a catalyst, without the accumulation of large volumes of biomass. However, currently there are no records of the use of the mer operon for the treatment of industrial waste or of other environments contaminated with mercury (von Canstein et al., 1999). Some experiments have been conducted in the form of a microcosmos (a glass apparatus with different chambers) used to perform environmental simulations (river, lake, etc.). In a central chamber the contaminated medium is treated with mercury-reducing bacteria. The concentration and form of mercury can be monitored in the different chambers. Mercury reduction from Hg2+ to Hg0 can reach a 95% rate when the Hg2+ in the first chamber (entry) is compared to that in the last one (exit), demonstrating the high biotechnological potential of mercury reduction by the mer operon (Saouter et al., 1994). Other studies, some of them conducted in our laboratory, have described mercury-reducing bacterial strains, with emphasis on Escherichia coli, obtained and genetically improved by means of mer operon cloning and by other recombinant DNA techniques (Hou et al., 1988; Nascimento et al., 1992a,b; Chen and Wilson, 1997; Cursino et al., 2000). MerA has been found to be active in yeast (Rensing et al., 1992) and plants (Rugh et al., 1996, 1998). Techniques to detect mercurial compounds in the environment using mechanical analysis procedures, such as atomic spectrophotometry (Omang, 1971) or cold-vapor atomic fluorescence detection (Bloom and Fitzgerald, 1988), have been developed. However, the preparation of samples is very laborious. An alternative will be the use of bacterial sensors. Bioassays can complement analytical chemical methods for the detection of biologically available mercury in environmental samples. Bacterial biosensors have been engineered to contain a report plasmid that carries gene fusions between the regulatory region of the mer operon (merR) and bacterial luminescence genes (lux-CDABE) that quantitatively respond to Hg2+ (Selifonova et al., 1993; Ramanathan et al., 1997; Rasmussena et al., 2000). REFERENCES Abelsohn, A., Gibson, B.L., Sanborn, M.D. and Weir, E. (2002). Identifying and managing adverse environmental health effects: 5. Persistent organic pollutants. Can. Med. Assoc. J. 166: 1549-1554. Bhriain, N.N. and Foster, T.J. (1986). Polypeptides specified by the mercuric resistance (mer) operon of plasmid R100. Gene 42: 323-330. Bloom, N. and Fitzgerald, W.F. (1988). Determination of volatile mercury species at the picogram level by low-temperature gas chromatography with cold-vapour atomic fluorescence detection. Anal. Chim. Acta 208: 151-161. Bogdanova, E.S., Bass, I.A., Minakhin, L.S., Petrova, M.A., Mindlin, S.Z., Volodin, A.A., Kalyaeva, E.S., Tiedje, J.M., Hobman, J.L., Brown, N.L. and Nikiforov, V.G. (1998). Horizontal spread of mer operons among gram-positive bacteria in natural environments. Microbiology 144: 609-620. Brown, N.L. (1985). Bacterial resistance to mercury-reductio ad absurdum? Trends Biochem. Sci. 10: 400-403. Brown, N.L., Misra, T.K., Winnie, J.N., Schmidt, A., Seiff, M. and Silver, S. (1986). The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol. Gen. Genet. 202: 143-151. Chang, J.S. and Hong, J. (1994). Biosorption of mercury by the inactivated cells of Pseudomonas aeriginosa PU21 (Rip64). Biotechnol. Bioeng. 44: 999-1006. Chen, S. and Wilson, D.B. (1997). Construction and characterization of Escherichia coli genetically engineered for bioremetiation of Hg2+-contaminated environments. Appl. Environ. Microbiol. 63: 2442-2445. Chen, W., Bruhlmann, F., Richins, R.D. and Mulchandani, A. (1999). Engineering of improved microbes and enzymes for bioremediation. Curr. Opin. Biotechnol. 10: 137-141. Cursino, L., Oberdá, S.M., Cecílio, R.V., Moreira, R.M., Chartone-Souza, E. and Nascimento, A.M.A. (1999). Mercury concentration in the sediment at different gold prospecting sites along the Carmo stream, Minas Gerais, Brazil, and frequency of resistant bacteria in the respective aquatic communities. Hydrobiologia 394: 5-12. Cursino, L., Mattos, S.V.M., Azevedo, V., Galarza, F., Bucker, D.H., Chartone-Souza, E. and Nascimento, A.M.A. (2000). Capacity of mercury volatilization by mer (from Escherichia coli) and glutathione S-transferase (from Schistosoma mansoni) genes cloned in Escherichia coli. Sci. Total Environ. 261: 109-113. Daher, V. (1999). No rastro do mercúrio. Ciênc. Hoje 26: 46-48. DeMagalhaes, M.E.A. and Tubino, M. (1995). A possible path for mercury in biological systems - the oxidation of metallic mercury by molecular oxygen in aqueous-solutions. Sci. Total Environ. 170: 229-239. Furukawa, K. and Tonomura, K. (1972). Mettalic mercury releasing enzyme in mercury-resistant Pseudomonas. Agric. Biol. Chem. 36: 217-226. Griffin, H.G., Foster, T.J., Silver, S. and Misra, T.K. (1987). Cloning and DNA sequence of the mercuric- and organomercurial-resistance determinats of plasmid pDU1358. Proc. Natl. Acad. Sci. USA 84: 3112-3116. Hamlett, N.V., Landale, E.C., Davis, B.H. and Summer, A.O. (1992). Roles of the Tn21 merT, merP, and merC gene products in mercury resistance and mercury binding. J. Bacteriol. 174: 6377-6385. Harada, M. (1978). Congenital Minamata disease: Intrauterine methylmercury poisoning. Teratology 18: 285-288. Hou, Y.M., Kim, R. and Kim, S.H. (1988). Expression of the mouse metallothionein-I gene in Escherichia coli: increased tolerance to heavy metals. Biochim. Biophys. Acta 951: 230-234. Huang, C.C., Narita, M., Yamagata, T. and Endo, G. (1999). Identification of three merB genes and characterization of a broad-spectrum mercury resistance module encoded by a class II transposon of Bacillus megaterium strain MB1. Gene 239: 361-366. Inoue, C., Sugawara, K. and Kusano, T. (1991). The merR regulatory gene in Thiobacillus ferrooxidans is spaced apart from the mer structural genes. Mol. Microbiol. 5: 2707-2718. Kalyaeva, E.S., Tiedjie, J.M., Hobman, J.L., Brown, N.L. and Nikiforov, V.G. (1998). Horizontal spread of mer operons among Gram-positive bacteria in natural environment. Microbiology 44: 609-620. Kholodii, G.Y., Yurieva, O.V., Lomovskaya, O.L., Gorlenko, Z.M., Mindlin, S.Z. and Nikiforov, V.G. (1993). Tn5053, a mercury resistance transposon with integron ends. J. Mol. Biol. 230: 1103-1107. Kiyono, M. and Pan-Hou, H. (1999). The merG gene product is involved in phenylmercury resistance in Pseudomonas strain K-62. J. Bacteriol. 81: 726-730. Komura, I. and Izaki, K. (1971). Mechanism of mercuric chloride resistance in microorganisms. I. Vaporization of a mercury compound from mercuric chloride by multiple drug resistance strain of Escherichia coli. J. Biochem. 70: 885-893. Lacerda, L.D. and Solomons, W. (1991). Mercury in the Amazon: A Chemical Time Bomb? CETEM/CNPq, Rio de Janeiro, RJ, Brazil, pp. 46. Laddaga, R.A., Chu, L., Misra, T.K. and Silver, S. (1987). Nucleotide sequence and expression of the mercurial-resistance operon from Staphylococcus aureus plasmid pI258. Proc. Natl. Acad. Sci. USA 84: 5106-5110. Langford, N. and Ferner, R. (1999). Toxicity of mercury. J. Hum. Hypertens. 13: 651-656. Liebert, C.A., Hall, R.M. and Summers, A.O. (1999). Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63: 507-522. McArthur, J.V. and Tuckfield, R.C. (2000). Spatial patterns in antibiotic resistance among stream bacteria: effects of industrial pollution. Appl. Environ. Microbiol. 66: 3722-3726. Misra, T.K. (1992). Bacterial resistances to inorganic mercury salts and organomercurials. Plasmid 25: 4-16. Misra, T.K., Brown, N.L., Fritzinger, D.C., Pridmore, R.D., Barnes, W.M., Haberstroh, L. and Silver, S. (1984). The mercuric-ion resistance operons of plasmid R100 and transposon Tn501: the beginning of the operon including the regulatory region and the first two structural genes. Proc. Natl. Acad. Sci. USA 81: 5975-5979. Mukhopadhyay, D.H., Yu, H., Nucifora, G. and Misra, T.K. (1991). Purification and functional characterization of MerD: a coregulator of the mercury resistance operon in Gram-negative bacteria. J. Biol. Chem. 266: 18538-18542. Munthe, J. and McElroy, W.J. (1992). Some aqueous reactions of potential importance in the atmospheric chemistry of mercury. Atmos. Environ. 26A: 553-557. Nascimento, A.M.A., Azevedo, M.O., Astolfi-Filho, S. and Chartone-Souza, E. (1992a). Cloning of the mercuric-ion resistance operon into Escherichia coli 5K using the mini-plasmid technique. Biotechnol. Tech. 6: 139-142. Nascimento, A.M.A., Azevedo, M.O., Astolfi-Filho, S. and Chartone-Souza, E. (1992b). Cloning of the mercuric-ion resistance operon of pBH100 into Escherichia coli 5K using pAT153 as vector. Rev. Microbiol. 23: 217-220. Nies, N.H. (1999). Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51: 730-750. Nishigaki, S. and Harada, M. (1975). Methylmercury and selenium in umbilical cords of inhabitants of Minamata area. Nature 258: 324-325. Nucifora, G., Silver, S. and Misra, T.K. (1990). Down regulation of the mercury resistance operon by the most promter-distal gene merD. Mol. Gen. Genet. 220: 69-72. Omang, S.H. (1971). Determination of mercury in natural waters and effluents by flameless atomic absorption spectrophotometry. Anal. Chim. Acta 53: 415-419. Osborn, A.M., Bruce, K.D., Strike, P. and Ritchie, D.A. (1997). Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 19: 239-262. Pan-Hou, H.S. and Imura, N. (1981). Role of hydrogen sulfide in mercury resistance determined by plasmid of Clostridium cochlearium T-2. N. Arch. Microbiol. 129: 49-52. Pan-Hou, H.S.K., Nishimoto, M. and Imura, N. (1981). Possible role of membrane proteins in mercury resistance of Enterobacter aerogenes. Arch. Microbiol. 130: 93-95. Pleijel, K. and Munthe, J. (1995). Modelling the atmospheric mercury cycle - 13687. Chemistry in fog droplets. Atmos. Environ. 29: 1441-1457. Ramanathan, S., Ensor, M. and Daunert, S. (1997). Bacterial biosensor for monitoring toxic metals. Trends Biotechnol. 15: 500-506. Radstrom, P., Skold, O., Swedberg, G., Flensburg, J., Roy, P.H. and Sundstrom, L. (1994). Transposon Tn5090 of the plasmid R751, which carries integron, is related to Tn7, Mu, and the retroelements. J. Bacteriol. 176: 3257-3268. Rasmussena, L.D., Sùrensena, S.J., Turnerb, R.R. and Barkay, T. (2000). Application of a mer-lux biosensor for estimating bioavailable mercury in soil. Soil Biol. Bioch. 32: 639-646. Reniero, D., Galli, E. and Barbieri, P. (1995). Cloning and comparison of mercury- and organomercurial-resistance determinants from a Pseudomonas stutzeri plasmid. Gene 166: 77-82. Rensing, C., Kues, U., Stahl, U., Nies, D.H. and Friedrich, B. (1992). Expression of bacterial mercuric ion reductase in Saccharomyces cerevisiae. J. Bacteriol. 174: 1288-1292. Ritter, J.A. and Bibbler, J.P. (1992). Removal of mercury from wastewater: large scale performance of an ion exchange process. Wat. Sci. Technol. 25: 165-172. Rugh, C.L., Wilde, H.D., Stack, N.M., Thompson, D.M., Summers, A.O. and Meagher, R.B. (1996). Mercuric ion reduction and resistance in transgenic Arabidopsis thaliana plants expressing a modified bacterial merA gene. Proc. Natl. Acad. Sci. USA 93: 3182-3187. Rugh, C.L., Senecoff, J.F., Meagher, R.B. and Merkle, S.A. (1998). Development of transgenic yellow poplar for mercury phytoremediation. Nat. Biotechnol. 16: 925-928. Sant’ana, Y.X., Chartone-Souza, E. and Ferreira, M.D. (1989). Drug resistance and colicinogeny of Salmonella typhimurium strains isolated from sewage-contaminated surface water and humans in Belo Horizonte, Brazil. Rev. Microbiol. 20: 41-49. Saouter, E., Turner, R. and Barkay, T. (1994). Microbial reduction of ionic mercury for the removal of mercury from contaminated environments. Ann. N. Y. Acad. Sci. 721: 423-427. Schottel, J.L. (1978). The mercuric and organomercurial detoxifying enzymes from a plasmid-bearing strain of Escherichia coli. J. Biol. Chem. 253: 4341-4349. Sedlmeier, R. and Altenbuchner, J. (1992). Cloning and DNA sequence analysis of the mercury resistance genes of Streptomyces lividans. Mol. Gen. Genet. 236: 76-85. Selifonova, O., Burlage, R. and Barkay, T. (1993). Bioluminescent sensors for detection of bioavailable mercury (II) in the environment. Appl. Environ. Microbiol. 59: 3083-3090. Silver, S. (1994). Exploiting heavy metal resistance systems in bioremediation. Res. Microbiol. 145: 61-67. Silver, S. (1996). Bacterial resistances to toxic metal ions - a review. Gene 179: 9-19. Silver, S. and Misra, T.K. (1984). Bacterial transformations of and resistances to heavy metals. Basic Life Sci. 28: 23-46. Silver, S. and Phung, L.T. (1996). Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50: 753-789. Summers, A.O. (1972). Mercury resistance in a plasmid-bearing strain of Escherichia coli. J. Bacteriol. 112: 1228-1236. Summers, A.O. (1986). Organization, expression and evolution of genes for mercury resistance. Annu. Rev. Microbiol. 40: 607-634. Summers, A.O. and Silver, S. (1972). Mercury resistance in a plasmid-bearing strain of Escherichia coli. J. Bacteriol. 112: 1228-1236. Summers, A.O. and Silver, S. (1978). Microbial transformations of metals. Annu. Rev. Microbiol. 32: 637-672. Trevors, J.T. (1986). Mercury methylation by bacteria. J. Basic Microbiol. 26: 499-504. Tuovinen, O.H. (1984). Mechanisms of microbial resistance and detoxification of mercury and organomercury compounds: physiological, biochemical and genetic analyses. Microbiol. Rev. 48: 95-124. von Canstein, H., Li, Y., Timmis, K.N., Deckwen, W.D. and Wagner-Dobler, I. (1999). Removal of mercury from chloralkali electrolysis wastewater by a mercury-resistant Pseudomonas putida strain. Appl. Environ. Microbiol. 65: 5279-5284. Walsh, C.T., Distefano, M.D., Moore, M.J., Shewchuk, L.M. and Verdine, G.L. (1988). Molecular basis of bacterial resistance to organomercurial and inorganic mercuric salts. FASEB J. 2: 124-130. Wang, Y., Mahler, I., Levinson, H.S. and Halvorson, H.O. (1987). Cloning and expression in Escherichia coli of chromosomal mercury resistance genes from a Bacillus sp. J. Bacteriol. 169: 4848-4851. Wang, Y., Moore, M., Levinson, H.S., Silver, S., Wash, C. and Mahler, I. (1989). Nucleotide sequence of a chromosomal mercury resistance determinant from a Bacillus sp. With broad-spectrum mercury resistance. J. Bacteriol. 171: 83-92. Weiss, A.A., Murphy, S.D. and Silver, S. (1977). Mercury and organomercurial resistance determined by plasmid in Staphylococcus aureus. J. Bacteriol. 132: 197-208. Wilson, J.R., Leang, C., Morby, A.P., Hobman, J.L. and Brown, N.L. (2000). MerF is a mercury transport protein: different structures but a common mechanism for mercuric ion transporters? FEBS Lett. 472: 78-82. |
|