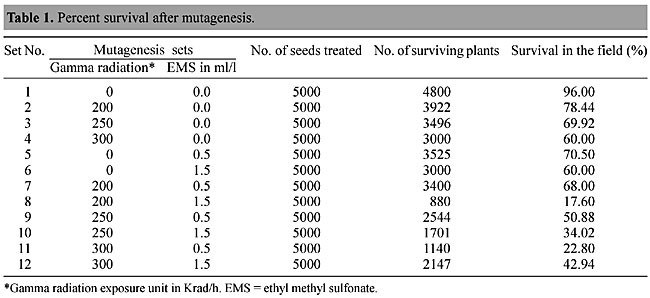
ABSTRACT. Over the last two decades, mutational techniques have become one of the most important tools available to progressive rice- breeding programs. In a mutation-breeding program initiated in 1999 at the Instituto Agronômico of Campinas, SP, Brazil, a rice line, IAC103, was selected for mutational studies with gamma radiation and ethyl methyl sulfonate mutagenesis, with the aim of developing a herbicide-resistant crop. After mutagenesis, surviving plants were exposed to glufosinate to check for herbicide resistance, which was examined up to the second generation. A detailed RAPD analysis was made of the resistant plants. Eighty Operon technology primers were tested and 10 were selected for a detailed study of RAPD markers that could tag herbicide resistance genes. Resistant and susceptible lines produced variation in the RAPD patterns and certain bands were found only in certain lines. These results suggest genetic ligation that will be confirmed through a genetic segregation study. Key words: Rice, Mutagenesis, Gamma radiation, Glufosinate, Ethyl methyl sulfonate, EMS, Segregation, Herbicide resistance INTRODUCTION Herbicide treatment of crops allows economically viable weed control and also provides cost-effective increases in the productivity of agricultural crops. Most of the herbicides currently in use combine good effectiveness with suitable production costs, are nontoxigenic and are rapidly biodegraded, hence they are “eco-friendly”. But some lack selectivity, therefore limiting their use to preemergence applications in the field. Breeding herbicide resistance into the crop is a new means to confer selectivity and enhance crop safety and production (Guttieri et al., 1992; Boutsalis and Powles, 1995; Hervieu and Vancheret, 1996). Herbicides generally affect processes that are unique to plants, e.g., photosynthesis or amino acid biosynthesis. These processes are shared by both weeds and crops. Therefore, developing herbicide-resistant crops is very difficult and a challenge to scientists, because every year a number of new herbicides are discovered. Generally two approaches to engineering herbicide resistance are used. In the first of these the target molecules in the cell are either rendered insensitive or are over-produced. In the second approach a metabolic pathway that degrades or detoxifies the herbicide is introduced into the plant (Tsaftaris, 1996). Phosphinothrin (PPT), commercially known as glufosinate or Round Up®, is an irreversible inhibitor of glutamine synthetase in plants and bacteria. Bialaphos, produced by Streptomyces hygroscopicus, consists of PPT and two alanine residues. When these residues are removed by peptidases the herbicidal component, PPT, is released. To prevent self-inhibition of growth, bialaphos-producing strains of S. hygroscopicus produce an acetyltransferase that inactivates PPT by acetylation. The bar gene that encodes acetylase has been introduced into many crops, including rice (Rathore et al., 1993; Jiang et al., 2000), to make transgenic herbicide-resistant crops. But this resistance can also be achieved by classical genetics and selection for over expression of glutamine synthetase, to make stable resistant lines instead of a plant modified by genetic engineering. We report the development of herbicide resistance by mutation and selection, using RAPD markers, for “Round Up” herbicide resistance in Brazilian rice cultivars. MATERIAL AND METHODS Rice varieties One rice line very susceptible to glufosinate, IAC 103, and 20 resistant lines (218-1,3-7; 219-1,3,5-10; 222-1 to 222-3; 197-1 to 197-2 and 165-3) were selected for analysis and designated as R1 to R21. These lines are continuously maintained in the greenhouses and research fields of the Instituto Agronômico of Campinas (IAC), Campinas, SP, Brazil, where they are periodically checked for herbicide resistance. Glufosinate application in the greenhouse The 21 rice lines were grown to the third or fourth leaf stage in the greenhouse and were tested for their response to glufosinate by spraying with a 1.0% (v/v) solution plus 0.1% (v/v) Tween 20. The actual concentration of PPT used was 480 mg/l. Resistant and susceptible plants were scored on the 15th day after treatment. DNA isolation and amplification For DNA isolation the methods of Sandhu et al. (2002) were followed. Fresh leaves (»2 g) were ground in liquid nitrogen and 300-400 mg of ground tissue transferred to polypropylene centrifuge tubes containing 15 ml of pH 8.0 extraction buffer (0.1 M Tris-HCl, 1.25 M NaCl, 0.02 M EDTA), 2% alkyltrimethylammonium bromide and 1% b-mercapto-ethanol. The mixture was slowly stirred for 90 min at 65ºC and an equal volume of 24:1 chloroform: isoamylalcohol added twice. The mixture was centrifuged at 10,000 g for 10 min and the supernatant transferred to a clean plastic tube containing 100 ml of a 10 mg/ml RNAse solution and incubated at 37ºC for 30 min, after which DNA pellets were obtained by adding 0.8 volumes of isopropanol. After washing with 70% ethanol, the DNA pellets were vacuum dried and dissolved in 200 ml of pH 8.0 TE buffer (10 mM Tris-HCl, 1 mM EDTA) and the quality and concentration of DNA fragments evaluated by electrophoresis in 0.8% agarose gels. This process was repeated for each of the 21 rice lines. PCR conditions PCR was carried out in a 25-ml reaction mixture containing 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, 10 ng template DNA, 1.0 mM primer, 100 mM of each dNTP and 1 unit of Taq polymerase. DNA was amplified in a Primus 96 Plus thermocycler (MWG-Biotech, Germany) at 96ºC for 4 min followed by 45 cycles of 1 min at 94ºC, 1 min at 35ºC, 1.3 min at 72ºC and a final stage of 7 min at 72ºC. The mixtures were maintained at 4ºC prior to analysis. For electrophoretic analysis 2.5 ml of 0.5% 1:2:1 bromophenol:blue:glycerol buffer was added and the amplification products loaded onto a 1.5% agarose gel in 1X TAE electrophoresis buffer. The gels were stained with ethidium bromide and photographed and analyzed using a Pharmacia Biotech gel documentation system. About 80 primers (Operon Technology, USA), OPF 1-20, OPJ 1-20, OPG 1-20 and OPK 1-20, were tested for polymorphism and differentiation in the 21 rice cultivar lines. Germination test of mature S1 seeds Seeds of self-pollinated S-0 mutated plants were designated as S1 seeds. In the first experiment, herbicide resistance was examined by germination of S1 seeds on RT medium (Murashige and Skoog, 1962) containing herbicide, RT medium-MS salts and eight vitamins, 20 g/l Phytagel (Sigma), pH 5.8. The actual concentrations of PPT used were 5 and 10 mg/l (Figure 12A-C). Germinated seeds were transferred to greenhouse (Figure 13A,B) and tested further for response to Round Up in a 1.0% (v/v) aqueous solution (Figure 13C). Mutational studies on rice lines About 5,000 seeds of Brazilian rice line IAC-103 (Oryza sativa var. Indica) were treated in 12 sets of mutagenic exposures (Table 1). These seeds were designated as the S-0 generation. Chemical mutagenesis was provoked by using ethyl methyl sulfonate (EMS; Sigma, USA), either alone or with gamma irradiation. Gamma radiation was applied with a Gammacell-220 machine (USA) at CENA, University of São Paulo, SP, Brasil. The doses used were 200, 250 and 300 Krad/h with a 60Co source, with or without exposure to the chemical mutagen, EMS, applied at two dosages: 0.5 ml/l or 1.5 ml/l. Standard methodology, as described by Camargo et al. (1996), Tisseli et al. (1996) and Tulmann et al. (2001) for gamma radiation and by Zhu et al. (1995) for EMS mutagenesis, was followed. After treatment all the seeds were germinated in a greenhouse and the percent survival was noted (Table 1). Surviving plants were transplanted to the IAC Rice Field, in Pindamonhangaba, SP, Brasil.
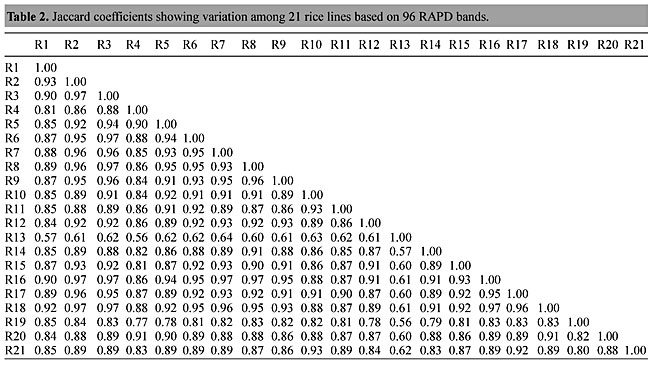
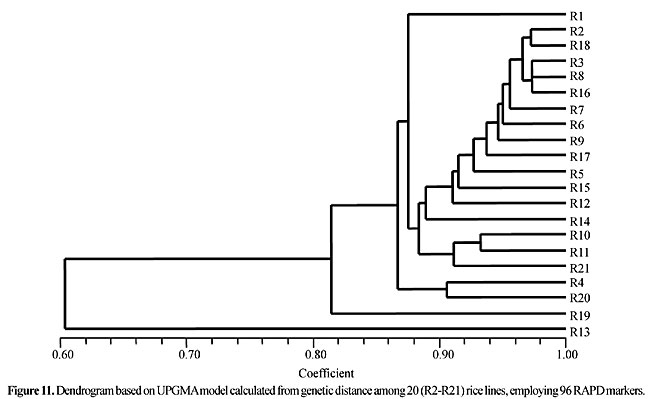
Three panicles per plant were harvested from these mutated plants and the seeds were designated as the S1 generation. Approximately 5,000 seeds from different sets of mutations were germinated in a greenhouse and then transplanted to the field, giving a total of 320 lines. They were exposed to the herbicide at the 4-5 leaf stage. The first application of glufosinate was 1.5 l/ha followed by a second application of 2 l/ha, 15 days after the first treatment. A third round was applied seven days after the second application. Surviving plants were grown until they produced seed, and three panicles per plant were harvested from each of them. These seeds were designated as the S2 generation. The S2 generation seeds were then germinated, subjected to herbicide resistance tests, and then planted in the rice field for later DNA analysis. Data analysis Only clearly amplified fragments were analyzed. Scores of 1 (present) or 0 (absent) were used to form a matrix. Simple matching coefficients (Sokal and Michener, 1958) were obtained to perform cluster (UPGMA) and principal coordinate analyses (PcoA). The variation set was represented by PcoA scores (99% total variation) based on RAPD markers. NTSYS-pc software (Rohlf, 1993) was used for the calculations. RESULTS AND DISCUSSION Screening of primers About 80 primers were tested on the 21 rice lines and among these, 13 were selected as suitable on the basis of good DNA amplification, with at least three sharp electrophoretic bands. Among these 13 primers, three, namely OPG18, OPG19 and OPF5, gave 100% polymorphism in all the 21 rice lines. The other 10 primers: OPK12, OPK19, OPF17, OPF19, OPJ7, OPJ9, OPJ16, OPJ17, OPG8 and OPG17 gave clear variations in the electrophoretic profile of the RAPD analysis of the 21 rice lines, and were thus considered to be suitable as RAPD markers for herbicide resistance in these lines. RAPD analysis In the comparison of resistant and susceptible lines, several DNA fragments produced variation in the RAPD patterns. In line R1 (IAC-103), K191050 (Figure 1) was found to be a strong marker for the susceptible line, but another marker, F171300 (Figure 2) was also found in this line, which corresponded to a marker of the resistant line, R19 (197-1). The variation in the R4 (218-4) line was the missing G17900 (Figure 3) band, which was found in the other lines. Another marker band not found in R4 was G17500, which corresponds to R19 (197-1), G17700 corresponded to R5 (218-5) and G171300 corresponded to R19 (197-1) and R20 (197-2); they could be treated as markers of resistance in these lines. The R5 (218-5) line varied at G17700, which also corresponds to R4 (218-4). 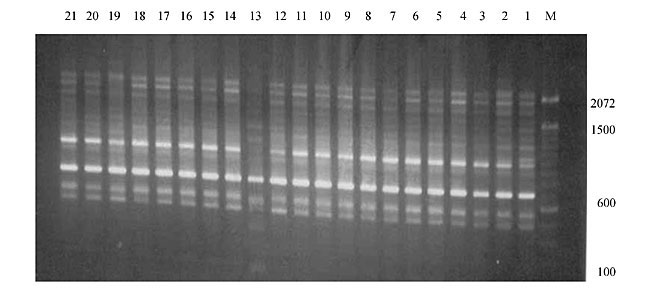 Figure 1. Ethidium bromide-stained electrophoretic profile of RAPD markers OPK19350, OPK192800 and OPK193000 in rice line R13, in comparison with 20 other rice lines. M = size marker. 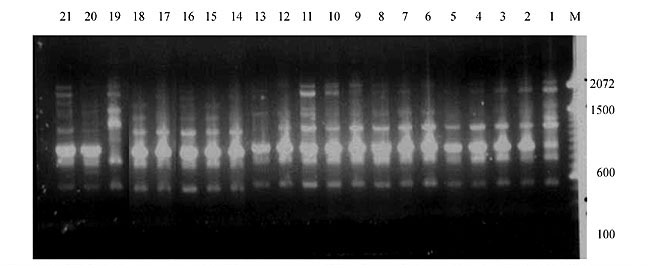 Figure 2. Ethidium bromide-stained electrophoretic profile of RAPD markers OPF17550 and OPF17600 in rice line R13, in comparison with 20 other rice lines. M is the 100-base pair DNA marker. 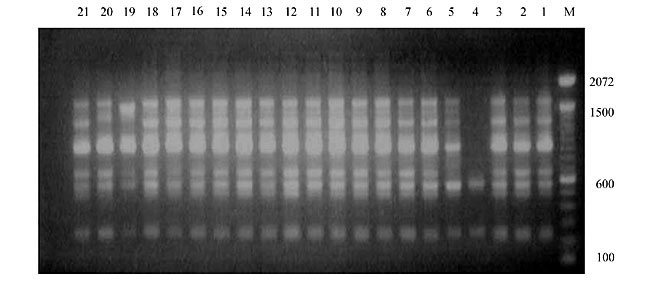 Figure 3. Ethidium bromide-stained electrophoretic profile of RAPD markers OPG17350 and OPG172800 in rice line R13, in comparison with 20 other rice lines. M = size marker. The largest variation in the resistant and susceptible lines was observed in the most susceptible lines, R1(IAC-103) and R13 (219-8). Seven fragments, OPJ7350 (Figure 4), OPJ172600 (Figure 5), OPK12550, OPK12600, OPK12900, OPK121100 (Figure 6) and OPK19400, were present in R13 and were not present in the susceptible line or in the other 19 resistant lines. At the same time, the following bands were missing in R13: K12650, K12700, K12850, K121000, K19950, K191700, K191900, K192300, OPJ72800, OPJ73000, OPJ9700, OPJ9800, OPJ91000 (Figure 7), OPJ16300, OPJ161200 (Figure 8), OPF171200, OPF171200, OPF19600 (Figure 9) and OPF19900. 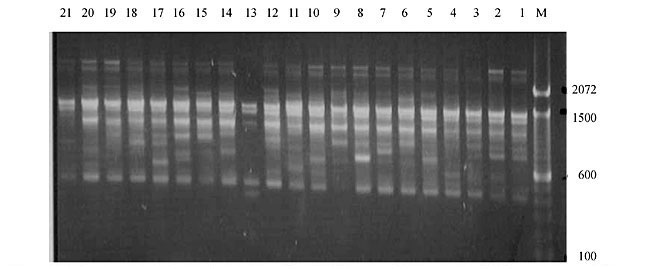 Figure 4. Ethidium bromide-stained electrophoretic profile of RAPD markers OPJ7550, OPJ7600, OPJ7900 and absence of J7650, J7850, J71000 in rice line R13, in comparison with other 20 rice lines. M = size marker. 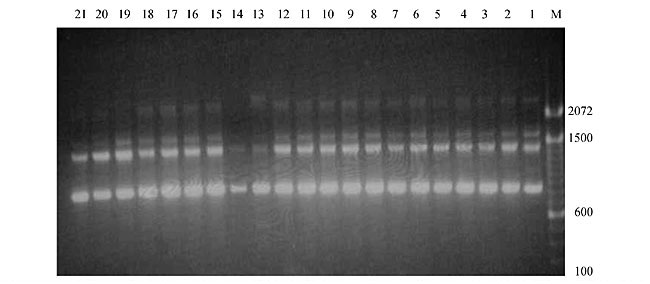 Figure 5. Ethidium bromide-stained electrophoretic profile of RAPD markers OPJ17350, OPJ172800 and OPJ173000 in rice line R13, in comparison with 20 other rice lines. M = size marker. 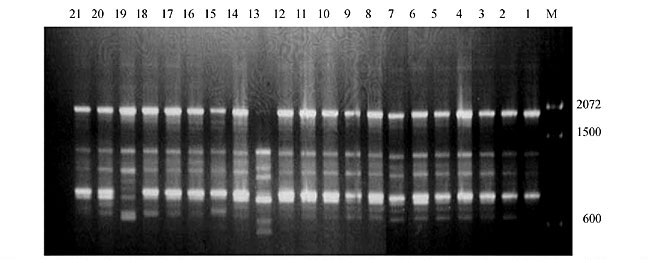 Figure 6. Ethidium bromide-stained electrophoretic profile of RAPD markers OPK12550, OPK12600 and OPK12900 and absence of K12650, K12850 and K121000 in rice line R13, in comparison with 20 other rice lines. M is the DNA marker (100-bp DNA ladder). 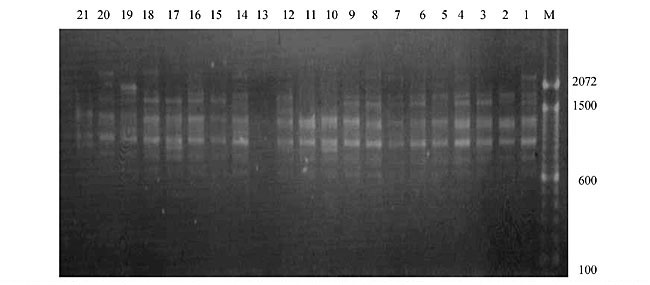 Figure 7. Ethidium bromide-stained electrophoretic profile of RAPD markers OPJ9350, OPJ92800 and OPJ93000 in rice line R13, in comparison with 20 other rice lines. M = size marker. 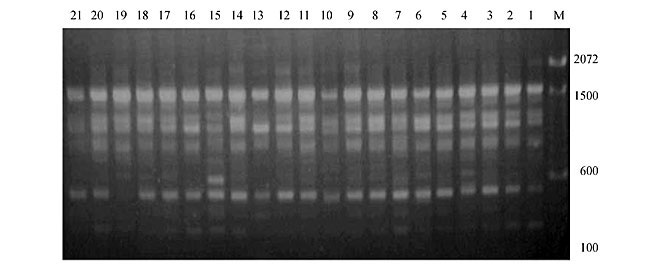 Figure 8. Ethidium bromide-stained electrophoretic profile of RAPD markers OPJ16550, OPJ16600 and OPJ16900 and absence of OPJ16650, OPJ16850, OPJ161000 in rice line R13, in comparison with 20 other rice lines. M = size marker. 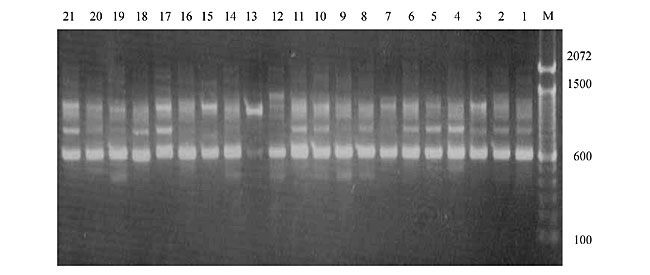 Figure 9. Ethidium bromide-stained electrophoretic profile of RAPD marker OPF19350 in rice line R19, in comparison with 20 other rice lines. M = size marker. Some variation was also observed in R14, as the OPJ171600 and OPJ172400 bands were absent. R19 exhibited variation as it had the OPF171300 band, which corresponded to susceptible line R1. OPG81800 (Figure 10) was present in R19, which corresponds to the resistance marker in R20. OPF17700 was found to be a separating marker for R20. 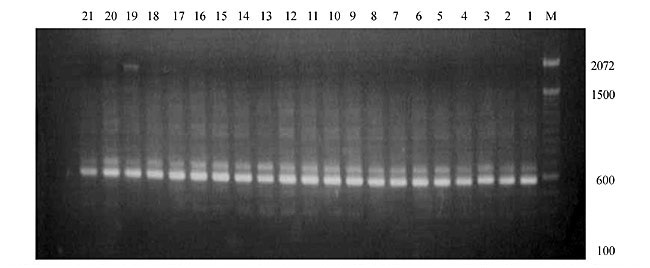 Figure 10. Ethidium bromide-stained electrophoretic profile of RAPD marker OPG81800 in rice line R19, in comparison with 20 other rice lines. M = size marker. The following DNA bands were absent in R19: OPK12850, OPK191900, OPG17500, OPG171300, while OPG81800 and OPF171300 were present. OPG81800 and OPF17700 were present in the R20 line, whereas OPG171300 was absent in R20. The R15 line showed variation, as OPJ16600 was present. The Jaccard coefficient results (Table 2 and Figure 11) also indicated that line R13 has the highest variability among the 21 lines.
Molecular markers are a more stable and informative alternative to isoenzymes. According to Cohen et al. (1991) and Colombo et al. (2000), these markers are more efficient for examining the genetic diversity of collections. Molecular markers have been used to study the genetic diversity of various species (Williams et al., 1993). Germination test of mature seeds from the S1 and S2 generations Glufosinate resistance was first demonstrated by the germination of S1 seeds in 5 mg/l and 10 mg/l glufosinate (Figure 12A-C). The plants were then transferred to a greenhouse (Figure 13A,B) and were found to be resistant to a 1.0% (v/v) aqueous solution of glufosinate. The resistance was also similar in the S2 generation in RT medium as well as in the greenhouse. This showed that herbicide resistance is transferred to the second generation. The first and second generation plants were also resistant in the field evaluation at the IAC rice field station (Figure 13D,E).     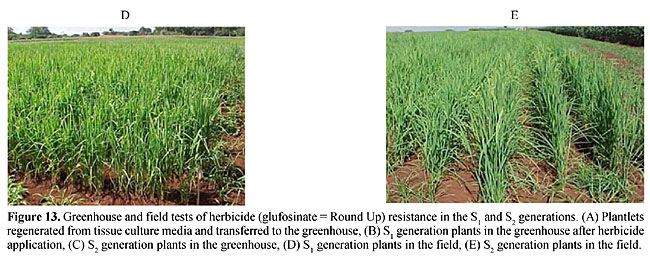 Over the last few decades mutational techniques have become among the most important tools available to progressive plant-breeding programs. Mutation and subsequent selection, with further breeding, has given stable and dominant rice lines with desired crop characters. Although we have not cloned the bar gene for PPT herbicide resistance in the rice, we were able to use classical genetics, mutation and selection on 20 rice lines to develop resistance against glufosinate. This resistance may be due to an over expression of glutamine synthetase, which would nullify the effect of PPT in the plants (Old and Primrose, 1998). Availability of a codominant RAPD marker for herbicide “PPT” resistance will be extremely useful for homologous resistance gene-pyramiding studies in these 21 rice lines to breed for herbicide resistance in Brazilian rice lines. ACKNOWLEDGMENTS S.S. Sandhu thanks the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasil) and TWAS, Italy, for financial support. The authors also thank CENA/USP, SP, Brazil, for providing the gamma irradiation facility. REFERENCES Boutsalis, P. and Powles, S.B. (1995). Inheritance and mechanism of resistance to herbicides inhibiting acetolactate synthase in Sonchus oleraceus L. Theor. Appl. Genet. 91: 242-247. Camargo, C.F. de O., Felicio, J.C., Ferreira, F.A.W.P., Pettinelli Junior, A., Tulmann Neto, A. and Ando, A. (1996). Mutational breeding of wheat in São Paulo State, Brasil. In: The Use of Mutation Techniques for Crop Improvement in Latin America (Ashri, A. and Bollich, C., eds.). IAEA, Vienna, Austria (IAEA-TECHDOC-859), pp. 95-111. Cohen, J.I., Alcorn, J.B. and Potter, C.S. (1991). Utilization and conservation of genetic resources: International Project for sustainable agriculture. Econ. Bot. 45: 190-199. Colombo, C., Gerard, S. and Charrier, A. (2000). Diversity within American Cassava germ plasm based on RAPD markers. Genet. Mol. Biol. 23: 189-199. Guttieri, M.J., Eberlein, C.V., Mallory Smith, C.A. and Thill, D.C. (1992). DNA-sequence variation in domain A of the acetolactate synthase genes of herbicide-resistant and herbicide-susceptible weed biotypes. Weed Sci. 40: 670-676. Hervieu, F. and Vancheret, H. (1996). A single amino acid change in acetolactate synthetase confers resistance to valine in tobacco. Mol. Gen. Genet. 251: 220-224. Jiang, J., Steve, D.L., Jianlin, W. and James, H.O. (2000). High efficiency transformation of U.S. rice lines from mature seed-derived calli and segregation of Glufosinate resistance under field conditions. Crop Sci. 40: 1729-1741. Murashige, T. and Skoog, F. (1962). A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol. Plant 15: 473-479. Old, R.W. and Primrose, S.B. (1998). The impact of recombinant DNA technology: the generation novelty. In: Principals of Gene Manipulation (Old, R.W. and Primrose, S.B., eds.). Blackwell Science, UK, pp. 369-407. Rathore, K.S., Chowdhury, V.K. and Hodges, T.K. (1993). Use of bar as a selectable marker gene and for the production of herbicide resistant rice plants from protoplast. Plant. Mol. Biol. 21: 871-884. Rohlf, F.J. (1993). NTSYS-pc. Numerical Taxonomy and Multivariate Analysis System. Version 1.80. Exter Software, Setauket, NY, USA. Sandhu, S.S., Colombo, C., Candido, B. and Walter, J.S. (2002). DNA tagging of blast resistant gene(s) in three Brazilian rice cultivars. Genet. Mol. Biol. (in press). Sokal, R. and Michener, C.D. (1958). A statistical method for evaluating systematic relationship. Univ. Kans. Sci. Bull. 38: 1409-1438. Tisseli, F.O., Azini, L.E., Bastos, C.R., Melode Castro, L.H.S. and Tulmann Neto, A. (1996). Increasing upland rice variability through induced mutation. In: The Use of Mutation Techniques for Crop Improvement in Latin America (Ashri, A. and Bollich, C., eds.). IAEA, Viena, Austria (IAEA-TECHDOC-859), pp. 17-22. Tsaftaris, A. (1996). The development of herbicide tolerant crops. Field Crops Res. 45: 115-123. Tulmann Neto, A., Alves, M.C., Camargo, C.E.O., Lopes de Castro, J. and Filho, W.P.F. (2001). New wheat genotype tolerant to aluminium toxicology obtained by mutation induction. Pesqui. Agropecu. Bras. 36: 61-70. Williams, J.G.K., Rafalski, J.A. and Tingey, S.V. (1993). Genetic analysis using RAPD markers. Meth. Enzymol. 218: 704-740. Zhu, B., Gu, A., Deng, X., Geng, Y. and Lu, Z. (1995). Effect of caffeine or EDTA post-treatment of EMS mutagenesis in soybean. Mutat. Res. 334: 157-159. |
|