ABSTRACT. As the pioneer among molecular cytogenetics techniques, fluorescence in situ hybridization (FISH) allows identification of specific sequences in a structurally preserved cell, in metaphase or interphase. This technique, based on the complementary double-stranded nature of DNA, hybridizes labeled specific DNA (probe). The probe, bound to the target, will be developed into a fluorescent signal. The fact that the signal can be detected clearly, even when fixed in interphase, improves the accuracy of the results, since in some cases it is extremely difficult to obtain mitotic samples. FISH is still used mostly in research, but there are diagnostic applications. New nomenclature is being developed in order to define many of the aberrations that were not distinguished before FISH. Prenatal diagnosis of aneuploidies and malignancies are promptly detected with FISH, which is very useful in critical cases. In some tumors, where chromosomal abnormalities are too complicated to classify manually, the technique of comparative genomic hybridization (CGH), a competitive FISH, allows examiners to determine complete or partial gain or loss of chromosomes. CGH results allow the classification of many tumor cell lines and along with other complementary techniques, like microdissection-FISH, PRINS, etc., increase the possibility of choosing an appropriate treatment for cancer patients. Key words: Molecular cytogenetics, Fluorescence in situ hybridization, Comparative genomic hybridization, Cancer patients
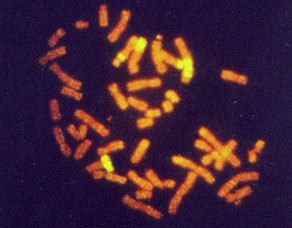
INTRODUCTION When the improvements in cytogenetics techniques seemed to have reached a plateau, a combination of cytogenetics and molecular biology gave rise to fluorescence in situ hybridization (FISH) opening many new opportunities in the field. The main reasons for this were: 1) It opens the possibility of obtaining cytogenetic results from interphase cells, impossible to achieve prior to the FISH technique. 2) Its colorful results are easy to interpret, explain and understand. 3) DNA is a very stable molecule, therefore archived material is suitable to be analyzed and paraffin-embedded tissues can be studied. 4) It can be combined with other cytogenetics and molecular techniques, complementing and improving the outcome of several other methods. Molecular cytogenetics are extensively used in many fields. Among these are: radiation biology, somatic cell genetics (cancer), prenatal and postnatal genetics, and preimplantational diagnosis. Some other fields are less explored, such as microbiology, ecology and evolution. Molecular cytogenetics techniques will have many new applications in probe synthesis for animal research models, and for specific diagnosis in economically important plant species, farm animals and pets. BRIEF DESCRIPTION OF APPLICATIONS Radiation biology FISH is one of the latest technologies used in radiation biology and it has brought many new insights to old problems such as basic research on aberration formation mechanisms. Initial chromosome damage and repair systems involve radiobiodosimetry (Cornforth et al., 1989; Mühlmann-Díaz and Bedford, 1993, 1994; Tucker et al., 1997; Edwards, 1997; Folle et al., 1998; Martinez-López et al., 1998) and predictive assays (Coco-Martin et al., 1994). Radiation-induced aberrations are one of the parameters analyzed after nuclear accidents. However, it is possible to estimate the risk the victims will have to confront and take preventive measures accordingly. Radiation-induced aberrations can be symmetrical or asymmetrical. The asymmetrical type aberrations (those which have an acentric fragment: dicentric, interstitial deletion, asymmetrical exchange, etc.) are lethal for the cells. Acentric fragments cannot be attached to the spindle in mitosis and usually they disappear, so cells carrying them will lack genetic information and are eliminated from the cell population a few generations post-irradiation. On the other hand, symmetrical type aberrations also decrease with post-irradiation time, but since they do not carry an acentric fragment they can survive as a clone in bone marrow, and therefore we can find them many years after exposure (Lucas et al., 1992). In whole body irradiation, we can estimate the exposure with a calculation of aberration frequency. FISH can be used for some of the chromosomes and then extrapolated to give the whole picture (Gebhart et al., 1996). Radiation is also used for medical purposes, and it is not uncommon to find patients who overreact to routine doses. On the other hand, some tumors are highly resistant to radiation therapy. Chromosome aberration frequencies, studied in vivo after irradiation of tumors, have been reported as a tool to distinguish tumor sensitivity or resistance to radiation. A noninvasive technique, that consists of aberration scoring in peripheral blood, is being developed in order to detect possible overreactions in patients that need radiotherapy. The same system is used to detect cancer-prone patients (Figure 1). For these studies, aberration scoring with plain Giemsa staining does not seem to have enough sensitivity. There have been some efforts to improve this method with FISH painting. This methodology has already been used in tumor cells (Coco-Martin et al., 1994).
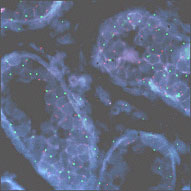
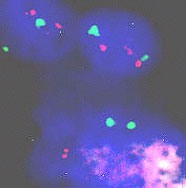
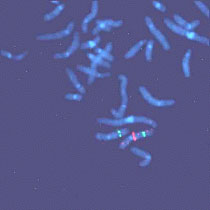
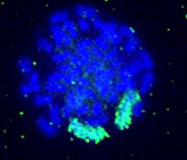
Somatic cell genetics In somatic cell genetics we can study features related to chromosome topology, nuclear structure and gene mapping (Sachs et al., 1993), as well as different malignancies and abnormalities related to cancer in hematological alterations and solid tumors (Siebert and Weber-Matthiesen, 1997; Cuneo et al., 1997). Chromosomal aberrations are found in cancer and tumor cell lines. Some of them are already characterized and correlated with specific syndromes (Tables 1 and 2) and some of them have yet to be associated with a clinical outcome. The continuous input of data and reports connecting clinical and biochemical data with chromosomal and genetic data will increase the list of known genes involved in each malignancy. The great advantage of the molecular cytogenetics techniques in these cases is the possibility to study interphase cells (Figure 2A). A good example is the amplification of the Her/2 neu oncogene (Figure 2B). About 30% of breast tumors (and other tumors) have amplification of this oncogene. These tumors have a bad prognosis, because they usually do not respond to the normal chemotherapeutic protocol and they are very aggressive. This oncogene codes for a membrane growth factor receptor. A humanized antibody against the specific receptor has recently been approved by the FDA and improves the condition of these patients considerably. Now they have an alternative. This is a typical example in which we can count on interphase cytogenetics of archived paraffin-embedded tissues.
Figure 2. Molecular cytogenetics techniques in interphase cells (A) and Her/2 neu oncogene (B). A. Paraffin-embedded tissue (60X): disgenesic gonads, X centromere (green), Y centromere ( red). B. Paraffin-embedded tissue (100X): Her/2 neu oncogene (red) amplified in breast cancer control hybridization (centromere 17 green). Chromosome triploidy involving chromosome 12 is a common feature of some leukemias. The fact that FISH with centromere probes allows us to count enough cells to make a statistical analysis of interphase cells makes the study of minimal residual disease (MRD) much faster and less demanding than by classical karyotyping.
Advanced molecular cytogenetics in neoplasias Interphase cytogenetics is indeed very useful when we know what we are looking for. But there is another problem in genetically unstable solid tumors. After the initial mutation some tumors become unstable and soon they have a wide range of different chromosomal aberrations. In any of these cases, comparative genomic hybridization (CGH) is a very useful tool. CGH is a special way to perform FISH (Figure 3A,B). It is a competition between
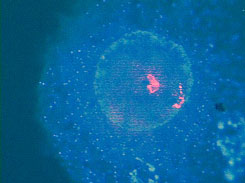
Figure 3B. Comparative genomic hybridization analysis examples. Left, Chromosome one: normal signal with green/red ratio about one. Right, Chromosome X with green/red ratio about 2.5, meaning supernumerary. normal cell DNA and suspected tumor DNA that allows the detection of deletions or amplifications that correspond to the entire tumor cell population. As with any methodology CGH complements other studies because by itself it will not detect balanced translocations. Nevertheless it is a very useful tool, because it allows us to recognize unbalanced aberrations, which are a common feature of the whole tumor (Hermsen et al., 1996). Homogenously stained regions (HSR) and double minutes are forms of DNA amplification that appear in some types of malignancies. The microdissection of chromosomes (Figure 4) and subsequent probe development (Xu et al., 1996) allow us to map a specific HSR to the DNA of origin. By making a probe from these microdissected areas we can use a normal cell as a target and find out where the amplified DNA comes from.
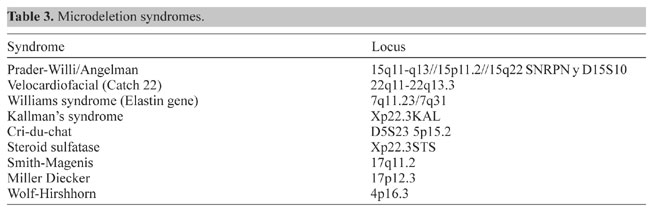
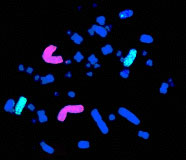
Figure 4. Microdissection procedure. Left, Selection of a chromosome piece for microdissection. Middle, Microneedle approaching the selected piece. Right, Area post-microdissection. Hereditary genetics Hereditary genetics includes syndromes passed on from the progenitors or that develop spontaneously during conception or embryogenesis. Prenatal testing. The fact that FISH allows us to perform interphase cytogenetics makes it suitable for very rapid prenatal diagnosis. Specific probes for the most common alterations can be used, giving a fast result while the regular cytogenetic results are still being processed. Even more important is that the probability of detecting any mosaicism for the probes that one is using is incremented, since metaphases and interphases are included in the scoring (Perandones et al., 1998). Postnatal, microdeletion syndromes. As mentioned before, there is no one methodology that can resolve all cases. Some syndromes and specific mutated loci can be detected or diagnosed by PCR, but in cases where there is a microdeletion involved, PCR is not useful since a quantitative PCR is very complicated. Specific locus FISH probes can give an answer. In these cases we have one chromosome in which a single locus is deleted, and the other is normal (Figure 5). Classical cytogenetics cannot detect these deletions, because they are microdeletions with only a few hundred base pairs (Table 3).
Preimplantational genetics. Some phenotypically normal persons carry balanced translocations or other types of aberrations. They usually have trouble having children or if they do, frequently their children have serious health problems. In other cases the abnormalities are spontaneous cases, but the parents are afraid to repeat the bad experience and seek prenatal testing for new pregnancies. In any case, waiting for the results causes considerable anxiety that can be minimized by using molecular cytogenetics techniques, such as FISH or PRINS. As FISH can be performed with a single cell, preimplantation diagnosis is possible (Figure 6). By knowing a priori what to look for, a combination of centromere probes can be used to detect aneuploidies (Coonen et al., 1998). When the female carries the translocation, pre-ICSI determination of the quality of the oocyte can be done with the polar body (Verlinsky et al., 1998). If the male carries the translocation, studies on the disjunction of the sperm can determine the probability that a single sperm is normal (Coco et al., 2000).
Figure 6. Human blastomere hybridized with total chromosome painting of chromosome 21: two chromosome 21 domains in red. Other less developed applications (Figure 7) but with just as promising a future are: Microbiology This area is perhaps the least developed but it further illustrates the multiple uses of molecular cytogenetics (Pillai, 1997). Diagnoses of the presence of certain viruses has been reported using probes for specific DNA of the infectious agent. Ecology Aberrations are an end point for genetic damage. Radiation by itself, or potentiated by other contaminants, can cause slight genetic damage (Ulsh et al., 2000). This might not affect phenotypic expression, but for example, translocation will decrease the pregnancy rate, and this in turn will slowly decrease the population growth of some species in the contaminated area. People might also be living in the surrounding areas, and be exposed to the same damage, affecting their life in the long run or even the next generation. Evolution During the speciation process in evolution some of the chromosome structures are conserved and some of them are interchanged with each other. The FISH technique has proved that related species not only have common genes but also common fragments of chromosomes (Mühlmann-Díaz et al., 2001; Wienberg and Stanyon, 1997). Agriculture and animal husbandry Chromosomal aberrations were first analyzed by banding techniques alone. Classical cytogenetics is based on species-specific bands to interpret the chromosomal exchanges that lead to aberrations. Training for these banding techniques is quite difficult and, every species has its own banding pattern, which the technician needs to learn. This is even more difficult for the many species for which banding does not work well and there is little background information in the literature to help with the classification of chromosomes. Once an appropriately labeled DNA probe is available, FISH becomes the technique of choice (Figure 7). That is why FISH has been so useful in different fields (Nkongolo et al., 1993; Goldammer et al., 1999). Figure 7. Applications of FISH probes in different species.
PAST, PRESENT AND FUTURE Molecular cytogenetics has gone a long way in a short time. Efficiency and sensitivity have been improved by combining methodologies. The FDA has already approved some of diagnostic testing techniques and more are under study for their approval. Still, many probes need to be developed, for more human specific genes and chromosome fragments, as well as for non-human species. Our group has been developing projects in this direction. REFERENCES Coco, R., Gallo, A., Mühlmann, M. and Provenzano, S. (2000). Estudio preconcepcional en un heterocigota de translocación t(1;4)(p32;q25). Análisis de segregación y aneuploidía por FISH doble y triple color. J. Bras. Reprod. Assist. 4: 51-59. Coco-Martin, J.M., Smeets, M.F.M.A., Poggensee, M., Mooren, E., Hofland, I., van den Brug, M., Ottenheim, C., Bartelink, H. and Begg, A.C. (1994). Use of fluorescence in situ hybridization to measure chromosome aberrations as a predictor of radiosensitivity in human tumour cells. Int. J. Radiat. Biol. 66: 297-307. Coonen, E., Hopman, A.H., Geraedts, J.P. and Ramaekers, F.C. (1998). Application of in situ hybridization techniques to study human preimplantation embryos: a review. Hum. Reprod. Update 4: 135-152. Cornforth, M.N., Meyne, J., Littlefield, L.G., Bailey, S.M. and Moyzis, R.K. (1989). Telomere staining of human chromosomes and the mechanism of radiation-induced dicentric formation. Radiat. Res. 120: 205-212. Cuneo, A., Bigoni, R., Roberti, M.G., Bardi, A., Balsamo, R., Piva, N. and Castoldi, G. (1997). Detection of numerical aberrations in hematologic neoplasias by fluorescence in situ hybridization. Haematologica 82: 85-90. Edwards, A.A. (1997). The use of chromosomal aberrations in human lymphocytes for biological dosimetry. Radiat. Res. 148: S39-S44. Folle, G.A., Martinez-Lopez, W., Boccardo, E. and Obe, G. (1998). Localization of chromosome breakpoints: implication of the chromatin structure and nuclear architecture. Mutat. Res. 404: 17-26. Gebhart, E., Neubauer, S., Schmitt, G., Birkenhake, S. and Dunst, J. (1996). Use of a three-color chromosome in situ suppression technique for the detection of past radiation exposure. Radiat. Res. 145: 47-52. Goldammer, T., Brunner, R.M., Kang’a, S., Hanotte, O. and Schwerin, M. (1999). Establishment of bovine chromosome region specific high resolution maps using microdissection. Anim. Biotechnol. 10: 113-118. Hermsen, M.A., Meijer, G.A., Baak, J.P., Joenje, H. and Walboomers, J.J. (1996). Comparative genomic hybridization: A new tool in cancer pathology. Hum. Pathol. 27: 342-349. Lucas, J.N., Awa, A., Starume, T., Poggensee, M., Kodama, Y., Nakano, M., Ohtaki, K., Weier, H.-U., Pinkel, D., Gray, J. and Littlefield, G. (1992). Rapid translocation frequency analysis in humans decades after exposure to ionizing radiation. Int. J. Radiat. Biol. 62: 53-63. Martinez-López, W., Boccardo, E.M., Folle, G.A., Porro, V. and Obe, G. (1998). Intrachromosomal localization of aberration breakpoints induced by neutron and gamma rays in Chinese hamster ovary cells. Radiat. Res. 150: 585-592. Mühlmann-Díaz, M.C. and Bedford, J.S. (1993). Breakage of human chromosomes 4,19 and Y in Go cells immediately after exposure to gamma-rays. Int. J. Radiat. Biol. 65: 165-173. Mühlmann-Díaz, M.C. and Bedford, J.S. (1994). A comparison of radiation-induced aberrations in human cells involving early and late replicating X chromosomes. In: Chromosomal Alterations: Origin and Significance (Obe, G. and Natarajan, T., eds.). Springer-Verlag, Berlin, Germany, 125-131. Mühlmann-Díaz, M.C., Ulsh, B.A., Whicker, F.W., Hinton, T.G., Congdon, J.D., Robinson, J.F. and Bedford, J.S. (2001). Conservation of chromosome 1 in turtles over 66 million years. Cytogenet. Cell Genet. 92: 139-143. Nkongolo, K.K., Lapitan, N.L.V., Quick, J.S. and Mühlmann, M.C. (1993). An optimized fluorescence in situ hybridization procedure for detecting rye chromosomes in wheat. Genome 36: 701-705. Perandones, C., Cerretini, R., Bozzo, W., Banares, V., Alba, L., Sala, A., Pivetta, O., Corach, D. and Mühlmann-Diaz, M.C. (1998). Diploid-triploid mosaicism in chorionic villus sampling (CVS): Clinical significance and prenatal assessment. Bio-Medicina 10: 425-429. Pillai, S.D. (1997). Rapid molecular detection of microbial pathogens: breakthroughs and challenges. Arch. Virol. 13 (Suppl.): 67-82. Sachs, R.K., Awa, A., Kodama, Y., Nakano, M., Ohtaki, K. and Lucas, J.N. (1993). Ratios of radiation-produced chromosome aberrations as indicators of large-scale DNA geometry during interphase. Radiat. Res. 133: 345-350. Siebert, R. and Weber-Matthiesen, K. (1997). Fluorescence in situ hybridization as a diagnostic tool in malignant lymphomas. Histochem. Cell. Biol. 108: 391-402. Tucker, J., Tawn, E., Holdsworth, D., Morris, S., Langlois, R., Ramsey, M., Kato, P., Boice Jr., J., Tarone, R. and Jensen, R. (1997). Biological dosimetry of radiation workers at the Sellafield nuclear facility. Radiat. Res. 148: 216-226. Ulsh, B.A., Mühlmann-Diaz, M.C., Whicker, F.W., Congdon, J.D., Hinton, T.G. and Bedford, J.S. (2000). Chromosome translocations in turtles: A biomarker in a sentinel animal for ecodosimetry. Radiat. Res. 153: 732-759. Verlinsky, Y., Cieslak, J., Ivakhnenko, V., Evsikov, S., Wolf, G., White, M., Lifchez, A., Kaplan, B., Moise, J., Valle, J., Ginsberg, N., Strom, C. and Kuliev, A. (1998). Preimplantation diagnosis of common aneuploidies by the first- and second-polar body FISH analysis. J. Assist. Reprod. Genet. 15: 285-289. Xu, J., Tyan, T., Cedrone, E., Savaraj, N. and Wang, N. (1996). Detection of 11q13 amplification as the origin of a homogeneously staining region in small cell lung cancer by chromosome microdissection. Genes Chromosomes Cancer 17: 172-178. Wienberg, J. and Stanyon, R. (1997). Comparative painting of mammalian chromosomes. Genet. Dev. 7: 784-791. |