|
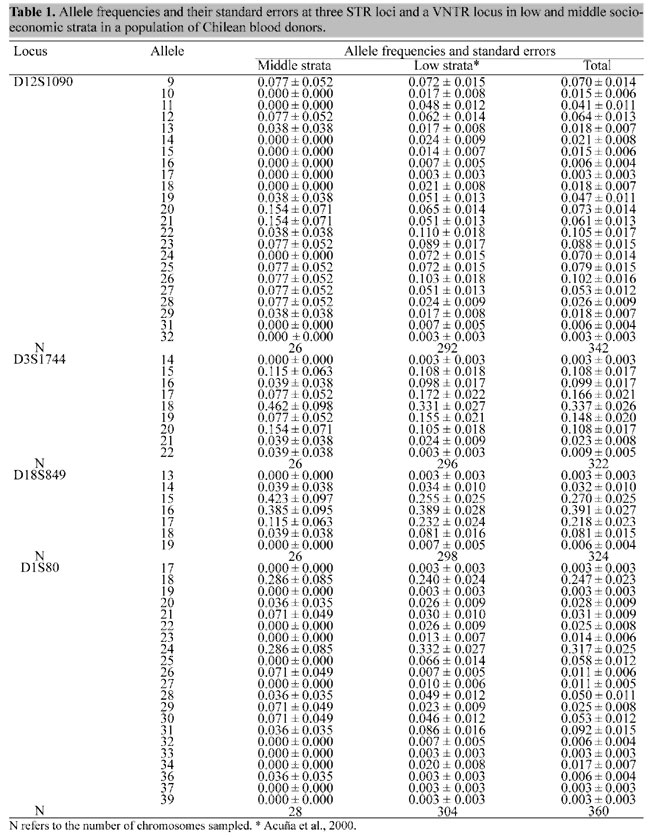
Frequency of the hypervariable DNA loci D18S849, D3S1744, D12S1090 and D1S80 in a mixed ancestry population of Chilean blood donors
ABSTRACT. Blood donors (N = 150) at San José Hospital (Santiago, Chile) were typed for one VNTR locus (D1S80) and three STR loci (D18S849, D3S1744, D12S1090). A questionnaire was used to determine the socioeconomic level of the donors, because it is known that some genetic markers (e.g., the ABO and Rh groups) are differentially distributed between different socioeconomic strata. This methodology revealed that two of the three socioeconomic strata distinguishable in Santiago were present in our sample of blood donors, with stratum II representing the middle strata and stratum III the low strata. Allele frequency was determined for each locus and socioeconomic stratum, and it was found that the allele distributions of each locus in socioeconomic strata II and III were statistically similar. All loci conformed to the Hardy-Weinberg law and there was no evidence for association between the alleles of the four loci, allelic frequencies being similar to those found in North American Hispanic populations. The results support the view that the analysis of these loci may have useful applications in population genetics as well as in identity tests. The inability to get or keep an rection in men.Sildenafil (tadapox for sale) is used to develop or maintainan erection. Know furosemide dosage?Key words: DNA polymorphisms, Chilean population, STR, VNTR, DNA typing, Forensic science
INTRODUCTION In Chile, allele frequencies for genetic markers are known to vary according to socioeconomic strata. For example, the A allele of the ABO blood group and the "d" allele are more frequent in the highest stratum, which has more genes of European origin, while native Amerindian genes are more prevalent in the lowest socioeconomic stratum (Valenzuela et al., 1987). The usefulness of genetic markers for identity testing and paternity analysis in Chile thus depends on knowing the allele frequencies in the different socioeconomic strata, but while the frequencies of the classical genetic markers are known (Valenzuela et al., 1987; Valenzuela, 1988) there have been no studies on socioeconomic class differences in allele frequencies using DNA markers. The present study describes the allele frequencies for one VNTR locus (D1S80) and three STR loci (D18S849, D3S1744, D12S1090) in an urban population of Northern Santiago, and compares these frequencies with data published in the literature and unpublished data from the Santiago forensic medicine service (Jorquera and Budowle, 1998). MATERIAL AND METHODS Blood samples (5 ml) were collected by venous puncture in tubes containing ACD from 150 unrelated blood donors at the San José Hospital blood bank whose catchment area is the Northern part of Santiago. DNA was extracted by an organic method described by Comey et al. (1994), primers for the D1S80 locus were those described by Kasai et al. (1990) and amplification and typing of the D1S80 were carried out by methods described by Budowle et al. (1995). The loci D18S849, D3S1744 and D12S1090 were analyzed using the Multiplex I kit (Lifecodes Corp.) and the alleles resolved by denaturing polyacrylamide gel electrophoresis according to the manufacturer's protocol. Each blood donor also responded to a socioeconomic classification questionnaire that was used to assign the donor to a socioeconomic strata based on power, prestige, occupation, educational level, income, life style, housing-district and other criteria (Sepúlveda, 1960). The Amerindian (Matson et al., 1967; Llop and Rothhammer, 1988) and Caucasian (Campillo, 1976) ethnic composition was estimated from the percentage of aboriginal admixture (Bernstein, 1931), using ABO and Rh (anti-D serum) systems as genetic markers. Allele frequencies at the four DNA loci were calculated by the gene counting method (Li, 1976) and the unbiased estimates of expected heterozygosity (H) as described by Edwards et al. (1992). Standard errors of allele frequencies were calculated as the square root of the variance of a binomial distribution. Possible divergence from Hardy-Weinberg equilibrium expectations was tested for by calculating the unbiased estimate of the expected homozygote/heterozygote frequencies (Nei and Roychoudhury, 1974; Nei, 1978; Chakraborty et al., 1988, 1991), using the likelihood ratio test (Chakraborty et al., 1991; Edwards et al., 1992; Weir, 1992), and an exact test (Guo and Thompson, 1992). An interclass correlation criterion (Karlin et al., 1981) for two-locus associations was used for detecting disequilibrium between the alleles at different loci (Chakraborty et al., 1993). The power of exclusion for each locus was determined according to the method of Weiner (1968). RESULTS AND DISCUSSION The questionnaire revealed two socioeconomic strata, stratum II (the middle socioeconomic strata) with about 23.7% aboriginal admixture and stratum III (low socioeconomic strata) with about 37.1% of aboriginal admixture. Allele frequencies and their standard errors were calculated from the genotypes of individuals for the two strata and the total population (Table 1). The allele distributions at each
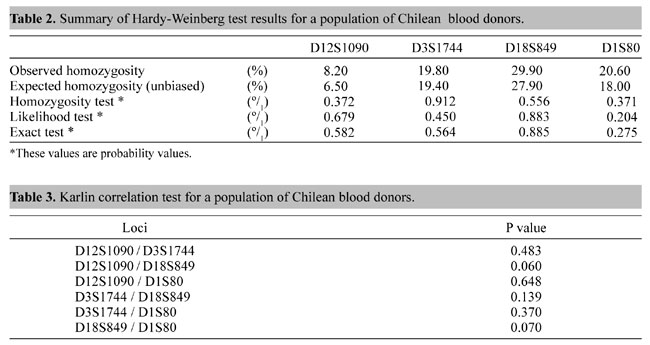
locus in strata II and III were statistically similar (P>0.05; c2 test). All loci were highly polymorphic and the blood donor database showed no evidence of departure from Hardy-Weinberg equilibrium (Table 2). There was no evidence of association between the loci using the interclass correlation test (Table 3).
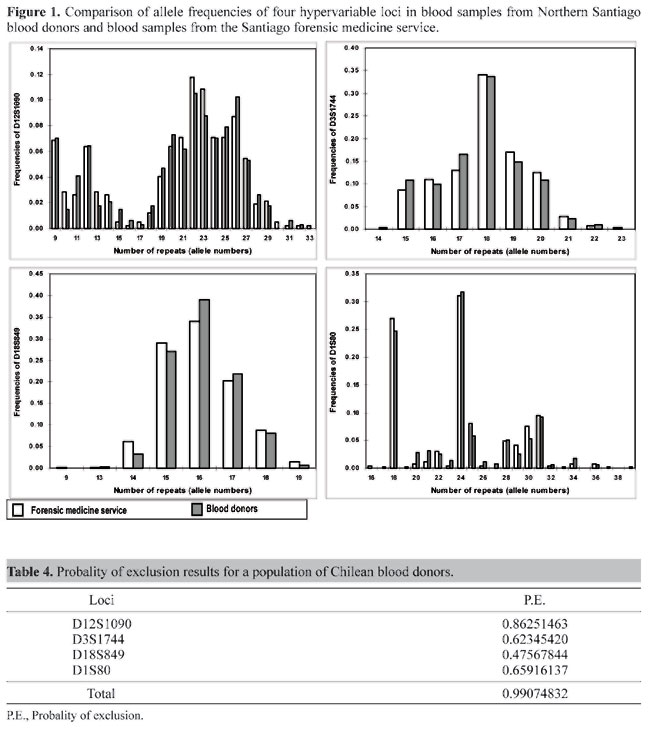
Comparisons of the distribution frequencies for all loci between the blood donor database and data from the Santiago forensic medicine service were made using the c² homogeneity test, but, as shown in Figure 1, no significant difference was observed between these two Chilean samples for any of the four loci. Figure 1 also compares the Chilean population and four ethnically different populations (US Black, US White, US Hispanic, US Oriental). Table 4 shows the probability of exclusion for paternity tests; the D12S1090 locus showed one of the highest powers of exclusion (0.8625) in our sample in relation to the other loci. The combined power of exclusion was 0.9908. Figures 2 and 3 show the frequencies of the different alleles of the four hypervariable DNA loci in different populations. After combining allele classes to avoid low expected values, a c² homogeneity test for allele frequency distribution between the populations was performed. Most alleles were observed in all populations, with the most frequent alleles usually not differing for each population. For the D18S849 locus the most frequent alleles for all five populations were 15, 16 and 17, with frequencies in the range of 13.6-41.7%. Our results for D18S849 in blood donors are very similar to those of US Hispanics (User's Manual; Lifecodes Corp.). The frequencies of different D3S1744 locus alleles in the five populations were similar, with allele 18 predominating. The c² test showed significant differences between the Chilean sample and US Black and US Oriental populations (User's Manual; Lifecodes Corp.). The allele distributions for the D12S1090 locus were bimodal and very similar for the five populations, but a c² homogeneity test for allele frequency distribution between the Chilean population and US Black, US White and US Oriental populations showed significant differences for this locus (User's Manual; Lifecodes Corp.).
Comparisons of D1S80 allele distribution between populations (Figure 2) using the c² test showed no significant differences between the Chilean population sample when compared with Southwestern US Hispanics and US Hispanics pooled (Zago et al., 1996; Huckenbeck et al., 1997). The research presented in this paper shows that the hypervariable DNA loci investigated are distinguishable from other Caucasian (Spanish), Black and Oriental populations, but that the D3S1744 locus is indistinguishable from the Caucasian population. All the loci studied are indistinguishable from USA Hispanic populations (Figures 2 and 3). This study provides the first database for DNA markers in low and middle socioeconomic strata in a Chilean population. The results presented indicate that the analysis of these loci may have useful applications in population genetics as well as in identity tests.
ACKNOWLEDGMENTS We thank Bruce Budowle for unselfish help with the statistics analysis, Ana Gladys Rivas for efficient laboratory work, and Lafayette Eaton for correcting the English and reviewing the genetic analyses. Ranjit Chakraborty made valuable comments on an earlier draft of the paper. Research supported by Fondecyt Project No. 1960405. REFERENCES Acuña, M., Jorquera, H., Armanet, L. and Cifuentes, L. (2000). Gene frequencies for four hypervariable DNA loci in a Chilean population of mixed ancestry. J. Forensic Sci. 45: 1160-1161. Bernstein, F. (1931). Die geographische Verteilung der Blutgruppen und ihre anthropologische Bedentung. In: Comitato Italiano per lo Studio dei Problemi Della Populazione. Instituto Poligrafico dello Stato, Rome, 227. Budowle, B., Baechtel, F.S., Smerick, J.B., Presley, K.W., Giusti, A.M., Parsons, G., Alevy, M.C. and Chakraborty, R. (1995). D1S80 population data in African Americans, Caucasians, Southeastern Hispanics, Southwestern Hispanics, and Orientals. J. Forensic Sci. 40: 38-44. Campillo, FL. (1976). Estudio de los grupos sanguíneos en la población Española. Comunicación de la Real Academia de Medicina. Tomo XCII de los Anales, Cuaderno tercero, Madrid. Chakraborty, R., Smouse, P.E. and Neel, J.V. (1988). Population amalgamation and genetic variation: observations on artificially agglomerated tribal populations of Central and South America. Am. J. Hum. Genet. 43: 709-725. Chakraborty, R., Fornage, M., Guegue, R. and Boerwinkle, E. (1991). Population genetics of hypervariable loci: analysis of PCR based VNTR polymorphism within a population. In: DNA Fingerprinting: Approaches and Applications (Burke, T., Dolf, G., Jeffreys, A.J. and Wolff, R., eds.). Birkhauser Verlag, Berlin, pp. 127-143. Chakraborty, R., Srinivasan, M.R. and Andrade, M. (1993). Intraclass and interclass correlations of allele sizes within and between loci in DNA typing data. Genetics 133: 411-419. Comey, C.T., Koons, B.W., Presley, K.W., Smerick, J.V., Sobieralski, C.A., Stanley, D.M. and Baechtel, F.S. (1994). DNA extraction strategies for amplified fragment length polymorphism analysis. J. Forensic Sci. 39: 1254-1269. Edwards, A., Hammond, H.A., Jin, L., Caskey, C.T. and Chakraborty, R. (1992). Genetic variation at five trimeric and tetrameric repeat loci in four human population groups. Genomics 12: 241-253. Guo, S.W. and Thompson, E.A. (1992). Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48: 361-372. Huckenbeck, W., Kuntze, K. and Scheil, H.G. (Eds.) (1997). The Distribution of the Human DNA-PCR Polymorphisms. Verlag Dr. Köster, Berlin, Germany, pp. 173-196. Jorquera, H. and Budowle, B. (1998). Chilean population data on ten PCR based loci. J. Forensic Sci. 43: 171-173. Karlin, S., Cameron, E.C. and Williams, P.T. (1981). Sibling and parent-offspring correlation estimation with variable family size. Proc. Natl. Acad. Sci. USA 78: 2664-2668. Kasai, K., Nakamura, Y. and White, R. (1990). Amplification of a variable number of tandem repeat (VNTR) locus (pMCT118) by the polymerase chain reaction (PCR) and its application to forensic science. J. Forensic Sci. 35: 1196-1200. Li, C.C. (1976). First Course in Population Genetics. Boxwood Press, Pacific Grove, CA, USA. Llop, E. and Rothhammer, F. (1988). A note on presence of blood groups A and B in pre-Colombian South America. Am. J. Phys. Anthropol. 75: 107-111. Matson, G.A., Sutton, H.E., Etcheverry, R., Swanson, J. and Robinson, A. (1967). Distribution of hereditary blood groups among Indians in South America IV. In Chile. Am. J. Phys. Anthropol. 27: 157-194. Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583-590. Nei, M. and Roychoudhury, A.K. (1974). Sampling variances of heterozygosity and genetic distance. Genetics 76: 379-390. Sepúlveda, O. (1960). Clasificación nacional jerarquizada de las ocupaciones expresadas en grandes estratos (ms). Facultad de Sociología, Universidad de Chile, Santiago, Chile. Valenzuela, Y.C. (1988). On sociogenetic clines. Ethol. Sociobiol. 9: 259-268. Valenzuela, Y.C., Acuña, M. and Harb, Z. (1987). Gradiente sociogenético en la población Chilena. Rev. Méd. Chile 115: 295-299. Weiner, A. (1968). Chances of proving non-paternity with a system determined by triple allelic co-dominant genes. Am. J. Hum. Genet. 20: 279-282. Weir, B.S. (1992). Independence of VNTR alleles defined by fixed bins. Genetics 130: 873-887. Zago, M.A., Silva, W.A., Tavella, M.H., Santos, S.E.B., Guerreiro, J.F. and Figueiredo, M.S. (1996). Interpopulational and intrapopulational genetic diversity of Amerindians as revealed by six variable number of tandem repeats. Hum. Hered. 46: 274-289. |